Abstract
Retroviral Gag proteins perform important functions in viral assembly, but are also involved in other steps in the viral life cycle. Conventional mutational analysis has yielded considerable information about domains essential for these functions, yet many regions of gag remain uncharacterized. We used genetic footprinting, a technique that permits the generation and simultaneous analysis of large numbers of mutations, to perform a near-saturation mutagenesis and functional analysis of 639 nucleotides in the gag region of Moloney murine leukemia virus. We report here the resulting functional map defined by eight footprints representing regions of Moloney murine leukemia virus gag, some previously uncharacterized, that are essential for replication. We found that significant portions of matrix and p12 proteins were tolerant of insertions, in contrast to the N-terminal half of capsid, which was not. We analyzed 30 mutants from our library by using conventional methods to validate the footprints. Six of these mutants were characterized in detail, identifying the precise stage at which their replication is blocked. In addition to providing the most comprehensive functional map of a retroviral gag gene, our study demonstrates the abundance of information that can be gleaned by genetic footprinting of viral sequences.
Retroviral Gag proteins primarily function in viral assembly. They also play important but less understood roles during viral entry into and release from cells. Gag precursors in Moloney murine leukemia virus (MoMLV) are proteolytically processed into four proteins: matrix (MA), p12, capsid (CA), and nucleocapsid (NC) (1). MA facilitates Gag transport to the cell surface in preparation for viral budding. p12 is responsible for nuclear transport of the viral DNA complex and later for virion release. CA is involved in assembly of the viral core and its disassembly after entry. NC binds the viral RNA genome and directs viral encapsidation by means of its Gag–Gag interaction domain (2). The specific regions of Gag that are important for these critical functions have not been fully defined. We adopted a mutational strategy in a large region of MoMLV Gag to identify regions essential for viral replication. Conventional methods of mutational analysis of viruses involve the individual isolation, storage, and characterization of mutants, because most mutants of interest are replication-defective and cannot be positively selected for. Such analysis is time-consuming and labor-intensive. Genetic footprinting (3) allows a comprehensive set of precisely defined mutations to be made and analyzed en masse to define essential regions in the sequence of interest. We have used this technique for high-resolution functional mapping of the bacterial supF gene (3) and HIV coreceptor CCR5 (4). It has also been used to analyze the 5′ end of HIV (5) and MoMLV envelope sequences (6).
We performed genetic footprinting of 639 nucleotides in the gag region of MoMLV. We report that large stretches of gag can tolerate 36-nt insertions. We define eight essential regions (footprints) of various lengths that do not tolerate insertions, generating a detailed functional map of MoMLV gag. We demonstrate a perfect correlation between these footprints and a conventional analysis of 30 randomly selected mutants, six of which are described in detail.
Materials and Methods
Creating a Library of Insertional Mutations in gag. A portion of gag between the restriction sites BsrGI and XhoI (nucleotides 1369–2014) in pNCA, the parent MoMLV plasmid (7), was subcloned into πAN13 to facilitate mutagenesis. MoMLV integrase was used to generate the library (3). The library contained 6.2 × 105 independent clones, averaging 400 insertional events per nucleotide. The insertion sequence was TGAAAGCTGCACGCGGCCGCGTGCAGCTTTCANNNN (N represents targetderived duplicated nucleotides). This library was subcloned into pNCA, yielding 7.5 × 106 proviral clones, with no loss in mutant diversity during subcloning. Sixty-one randomly selected mutations were sequenced, of which 52 (85%) had the predicted size and sequence. Fifteen percent had a frameshift mutation due to a nonstandard cut made by integrase, resulting in either a 3- or a 5-bp repeat flanking the insertion, instead of the canonical 4-bp repeat.
Cell Transfection and Infection. 293T cells and Rat2 cells were maintained in DMEM supplemented with 10% and 5% FBS, respectively, with penicillin (100 units/ml) and streptomycin (100 μg/ml). Lipofectamine Plus (Life Technologies, Grand Island, NY) was used to transfect 293T cells with the library. The resulting pool of mutant virions was harvested within 24 h, titered by using an assay for reverse transcriptase (RT) (8), and used to infect Rat2 cells at low (one virus per 100 cells) multiplicity of infection (moi). Mutations that did not affect viral replication resulted in release of progeny virus into the medium, which was harvested and used to infect more cells at low moi. Infections with individual mutants were performed as described (8, 9).
Footprinting. PCR analysis of gag mutants was performed as described (3), using a primer complementary to a region within the insertion (5′-GGCCGCGTGCAGCTTTCA) and a second radiolabeled primer that hybridized to a site within gag [1311U, 5′-CCTACATCGTGACCTGGGAAGC; 1352U, 5′-CCCTGGGTCAAGCCCTTTGTAC; 1761L, 5′-TCCAGTTGTAAAGGTCAGAAGAGG; 2026L, 5′-CCTACCTGCCTGGGTGGTGTAATCC; 1608U, 5′-GAGAAGCGACCCCTGCGGGAG; and 2134L, 5′-ATTGGGCCCTTGTGTTATTCCT]. Low molecular weight DNA from infected cells (10) was used as template in the PCR. PCR products were run on denaturing polyacrylamide gels.
Virus Purification and Analysis of Viral Proteins. Forty-eight hours after transfection of 293T cells with wild-type or mutant proviral DNAs, supernatant was collected. Virions were pelleted and cells were lysed as described (3, 8, 11). Virions and cell lysates were immunoblotted by using goat anti-CA serum (National Cancer Institute serum 79S-804, from S. P. Goff, Columbia University) diluted 1:5,000.
PCR Analysis of Specific Steps in Viral Life Cycle. We analyzed low molecular weight DNA from infected cells by using the following primers. For minus-strand strong-stop DNA, we used LTR2, 5′-AGTCCTCCGATTGACTGAG, and ss-as, 5′-CGGGTAGTCAATCACTCAG (9). For gag, we used the primers 5′-GGCCGCGTGCAGCTTTCA and 5′-TTTCTTCCGGGGTTTCTCGTTT. For mtDNA, we used the primers 5′-GTTAATGTAGCTTATAATAAAGC and 5′-GTTTAGGGCTAAGCATAGTGGG (12). Thermocycling conditions were as described (3), except for annealing temperatures, which were 57°C for minus-strong stop and mtDNAs, 63°C for gag, and 50°C for LTRs.
Results
Creating a Library of Insertional Mutations in gag. Genetic footprinting involves creating a large set of mutations in a gene and subjecting them to selection for function (Fig. 1). MoMLV integrase was used to make a library of near-random insertions into a region of gag spanning the coding regions for the C-terminal half of MA, all of p12, and an N-terminal portion of CA. Each mutant contains a single insertion of a defined 36-nt sequence, encoding one of three amino acid sequences depending on the reading frame (LESCTRPRAAFT in frame 1, LKAARGRVQLSL in frame 2, and a stop codon in frame 3).
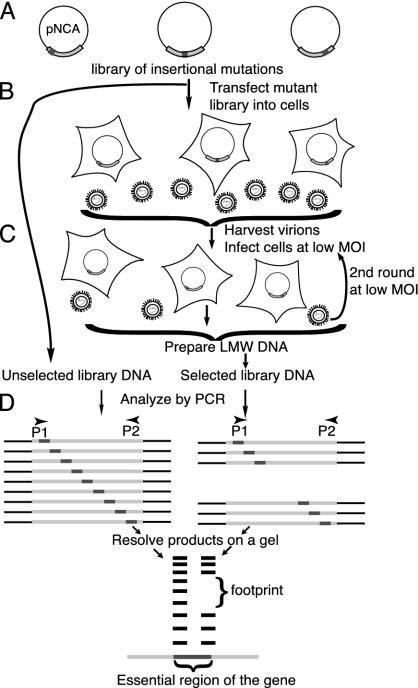
Fig. 1.
Scheme for generating and analyzing mutations. (A) Library of mutations. Members of the library contained a single insertion at a random position in gag within pNCA, the proviral construct. (B and C) Selection for gag function. 293T cells were transfected with the library. Virus was harvested from these cells and used to infect Rat2 cells at low moi. Virions were harvested and used to infect a second set of Rat2 cells at low moi, thus completing at least one cycle of replication. Low molecular weight (LMW) DNA prepared from infected Rat2 cells constituted selected DNA. (D) Analysis by PCR. Unselected DNA and selected DNA served as templates. Primer P1 corresponded to the insertion; P2 primed from defined sites in gag. PCR products run on a sequencing gel produced a ladder of bands, each band corresponding to at least one independent mutant. Mutants that failed selection gave rise to footprints. Comparing the sizes of missing bands with bands of known size allowed precise mapping of essential regions in gag.
Viral Replication as Selection for gag Function. The library was introduced into 293T cells (Fig. 1B), chosen for their high transfection efficiencies (60–80%) and their ability to support late events in the viral life cycle, such as transcription, translation, assembly, and release of virions. These properties allowed us to generate sufficient virions from the library. 293Ts lack the receptor for MoMLV, thus viral entry and subsequent steps of infection were studied by infecting Rat2 cells with virus obtained from 293Ts (Fig. 1C). Low moi (0.01 virion per cell) prevented functionally defective virions from complementing each other to produce viable virus. The ability of the virions to complete their life cycles depended on functional Gag proteins, allowing viral replication to be a selection for gag function. Virions in the supernatant were used to infect a second set of Rat2 cells, again at low moi. Low molecular weight DNA (10) from these cells contained DNA from only those viruses that were able to complete the entire replication cycle. This selected DNA, along with unselected DNA (made from the library before selection), was analyzed by PCR (Fig. 1D).
Footprinting. PCR was performed on unselected and selected DNA (Fig. 1D). A primer complementary to a site within gag and another complementary to the insertion were used, resulting in products of different lengths that separated at single base resolution on polyacrylamide gels. Mutants that failed selection did not give rise to a PCR product, resulting in footprints representing essential regions of the gene. The precise location and sequence of mutations that disrupt gene function could be determined without the isolation and sequencing of individual mutants.
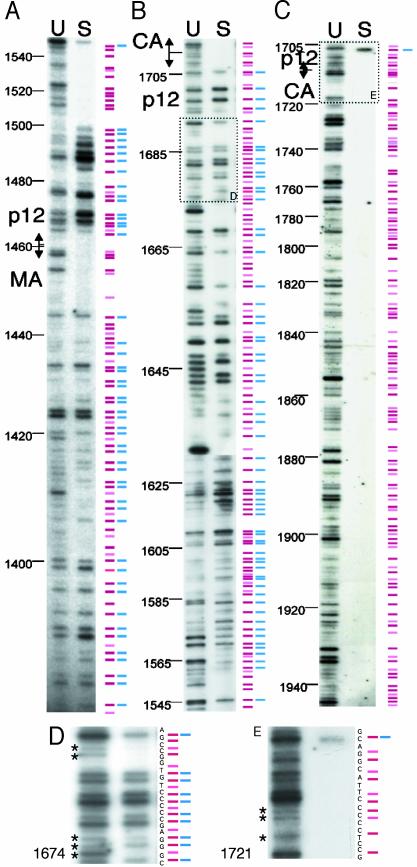
A representative set of gels from such an analysis is shown in Fig. 2, containing footprints from the C-terminal half of MA and an N-terminal portion of p12 (A), the rest of p12 and the p12/CA border (B), and the N-terminal half of CA (C). “U” lanes contain PCR products from the unselected library, each band representing at least one insertion at a specific site. “S” lanes contain PCR products from the selected pool and lack many bands seen in the U lanes, forming footprints.
Fig. 2.
Genetic footprinting of MoMLV gag. Footprinting analyses of gag spanning nucleotides 1380–1946. (A) C terminus of MA and part of p12 (nucleotides 1380–1550). (B) Rest of p12 and p12/CA border (nucleotides 1543–1715). (C) N terminus of CA (nucleotides 1705–1946). DNA from the unselected and selected libraries was used as a template for PCR. A primer complementary to the insertion was paired with one of many radiolabeled primers complementary to defined sites in gag. These sites lie within and just outside the region of mutagenesis. Lanes U contain PCR products made from unselected libraries, and lanes S contain products from selected libraries. Products were run adjacent to sequencing reactions (not shown) to allow the precise sizing of bands. A diagrammatic representation of bands is shown alongside the lanes. Red bars, read-through mutations; pink bars, mutants with stop codons; blue bars, viable mutants; and absence of blue bars, footprints. This figure is the result of several PCRs, with many primers, priming in both directions. The gels shown are representative of those used in the complete analysis. D and E show longer exposures of the areas outlined by dashed boxes in B and C. Bands not seen in shorter exposures are marked by asterisks. The nucleotide sequence of the relevant region of gag is depicted next to the bands to indicate the exact location of the corresponding insertions.
A diagram of our footprinting results is shown alongside lanes U and S, with red bars representing read-through insertions and pink bars representing insertions encoding stop codons, thus denoting truncations. Not surprisingly, none of the truncations was replication-competent. Viable viruses resulted in bands in lane S and are denoted by blue bars alongside red bars. The absence of a blue bar indicates that an insertion at that site is not compatible with viral replication. Although the relative intensity of PCR products from the selected and unselected pools could distinguish subtle quantitative defects in a mutation, blue bars are used to mark all mutations that survived selection, regardless of their fitness.
The pattern of bands seen in the U lanes indicates that integration events that created the library were sufficiently random to represent a large number of insertions in gag. However, the frequency of integration varied among the different sites, resulting in variable intensity of the corresponding bands. The diagram in Fig. 2 is derived from several gels, and, whereas it usually matches the bands from the representative gels shown, it is not an exact match for rare insertions. Optimal visualization of bands from these rare insertions required prolonged autoradiographic exposures, at which point it was difficult to resolve the more prevalent insertions. Prolonged exposures of boxed regions from Fig. 2 B and C are shown in Fig. 2 D and E, where less prevalent insertions (marked by asterisks) are better visualized. Many primer pairs were used in the PCR analysis. It was confirmed that each band resulting from a PCR was related to a specific mutant, and its intensity remained constant regardless of the primer used in the analysis (not shown). When a run of sequential bands was seen in lane U, every third band in lane S was absent. These absent bands correspond to the pink bars and are due to a stop codon in one reading frame. They have little relevance to our analysis and were disregarded.
PCR products of known sizes generated from sequenced mutations, in conjunction with DNA-sequencing reactions (not shown), enabled us to assign each insertion to an exact internucleotide position. Determining the position of each band and footprint led to a detailed functional map of gag (Fig. 3). A review of this map reveals large variations in tolerance to insertions seen in different regions of gag. Whereas several regions in MA and p12 tolerate insertions, no region in CA does. Footprints in each region are discussed below.
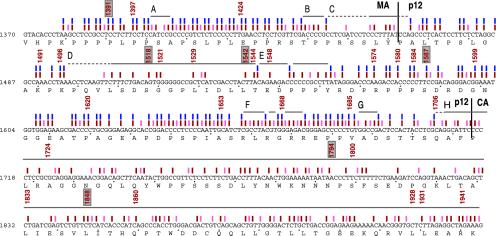
Fig. 3.
Functional map of gag. The nucleotide sequence of the mutagenized region of gag is shown, with the amino acid sequence directly below. Colored bars are described in the legend to Fig. 2. Footprints whose extent was greater than three amino acids in length are marked by horizontal lines and assigned a letter code (A–H). Where the boundaries of a footprint are ambiguous because of a paucity of insertions in that region, the horizontal lines are dashed. The location of the 30 mutants analyzed by conventional methods are denoted by the nucleotide number immediately 5′ to the insertion. Mutants subjected to further detailed analysis (see Fig. 5) have a box drawn around their nucleotide number.
Footprints in the C-Terminal Region of MA. The sequence corresponding to the C-terminal portion of MA included in this mutagenesis was almost saturated with insertional mutations. Most of these resulted in replication-competent virions (Figs. 2 A and 3). However, three small regions (A, B, and C) of 4 nt or more appear to be essential for viral replication. No function has been ascribed to regions A (nucleotides 1402–1406) or B (nucleotides 1440–1444) yet. Region C, at the MA–p12 border (nucleotides 1446–1465), extends into p12, possibly up to nucleotide 1465. A paucity of insertions in this region prevented us from accurately defining the 3′ margin of this footprint. Some of this region might be essential for the proteolytic cleavage between MA and p12, indicating that this cleavage is necessary for viral replication.
Footprints in p12. The p12 region had footprints of various sizes (Figs. 2 A and B and 3). The first 34 nucleotides of p12 were tolerant of insertions. Region D (nucleotides 1498–1544), although not a favored target for insertion, represented the largest footprint in p12. We performed a detailed analysis of two mutants within this large footprint (p12-1518 and p12-1542) and one outside this region (p12-1587; see below). We found that for both mutants in this region, when proviral DNA was introduced into cells, near wild-type levels of virions were assembled and released, indicating that late steps in the life cycle were not likely to be affected. However, mutant p12-1518 appeared to be blocked at early stages of viral entry, because when it was used to infect cells, no reverse transcription was detected. Mutant p12-1542 was capable of reverse transcription but its viral DNA was not seen to enter the nucleus (data below). Thus, this large footprint appears to contain features essential for the early stages of viral entry, most likely nucleocapsid disassembly, and for transport of the viral DNA complex into the nucleus (13). Adjacent to this footprint was region E, which contained only one viable insertion within a span of 16 nucleotides (1546–1562). The PPPY late-assembly domain, known to mediate viral budding (14), lies within this region, flanked by a viable mutation on each side. Note the “Y” in the PPPY domain is not included in the footprint because the insertion sequence at that location re-creates the Y codon. The C-terminal portion of p12 from nucleotides 1563 to 1706 was a frequent target for insertions and was largely tolerant of them. Mutant p12-1587, located within this region, was indistinguishable from wild-type virus in our analysis. This region also contained two footprints. Footprint F (1659–1673) contains three viable mutations and also contains arginine residues known to be essential for early stages of replication (15). Footprint G (1687–1691) has no previously identified function. A large footprint starting at the p12–CA border, footprint H (1707–downstream), indicates that proteolytic processing between p12 and CA is essential for replication.
Footprints in the N-Terminal Region of CA. Footprint H spans the entire sequence coding for the N-terminal half of CA, implying that extensive portions of CA are essential for some step in the viral life cycle. This region has not been mutagenized to any significant extent before. Other parts of CA that have been mutagenized, are also largely intolerant of mutations, both insertions and point mutations (16–19). A stepwise analysis of two randomly selected CA mutants is described below.
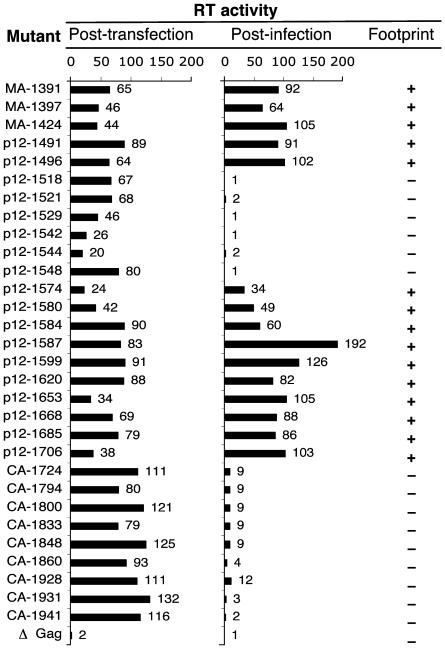
Validation of Footprints by Conventional Methods. We chose 45 mutants at random from the library for conventional analysis. Fifteen of these had stop codons and were nonviable. Map locations of the remaining 30 mutations are shown in Fig. 3, and results from their conventional analysis are shown in Fig. 4. Mutant proviral DNAs were individually introduced into 293T cells. Cell supernatants were assayed for virions by measuring RT activity. The presence of RT activity in the supernatant indicated that the mutants were capable of late steps in the viral life cycle, such as viral RNA and protein synthesis and virion assembly and release (see Fig. 4, posttransfection). The RT activity of wild-type virus was 100% and that of a mutant containing stop codon in gag was 2%. Each of the 30 mutants resulted in some RT activity in the supernatant, indicating that late steps in the viral life cycle remain largely unimpaired for most insertions in this region. The supernatants were next used to infect Rat2 cells, and RT activity was assayed in the Rat2 cell supernatants (Fig. 4, postinfection). The presence of RT activity after infection indicated that these mutants were able to complete the replication cycle. Each of these viable mutants lay outside the footprints, indicating a perfect correlation between footprinting and conventional analysis.
Fig. 4.
Validation of footprinting analysis by conventional methods. Mutant proviral DNAs were individually introduced into 293T cells. Medium was harvested 48 h later (posttransfection) and assayed for RT activity (indicating virion assembly and release). This supernatant was used to infect Rat2 cells, and RT activity was assayed 48 h later (postinfection). Values for RT activity are relative to wild type, which was set at 100%. The right-hand column shows whether these mutants were located within (–) or outside (+) footprints.
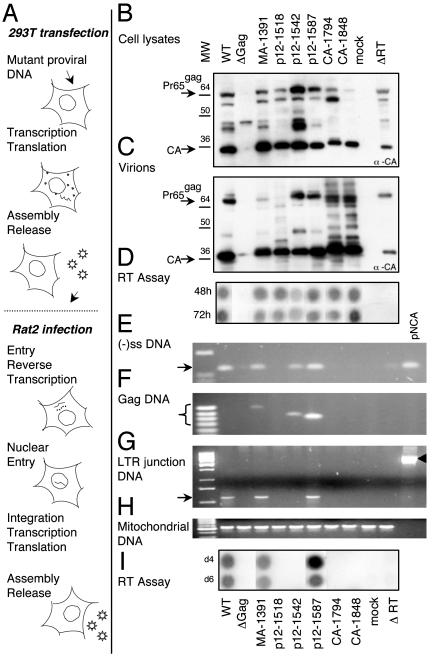
Detailed Analysis of Six Mutants. Six mutants were selected at random, some within footprints (p12-1518, p12-1542, CA-1794, and CA-1848) and others outside (MA-1391 and p12-1587). These mutants were followed through specific stages of the life cycle (Fig. 5A). Proviral DNA corresponding to each mutant was individually introduced into 293T cells to study late steps in the viral life cycle and to obtain virions for subsequent studies on viral entry. A truncation mutant in gag (Δgag) and a deletion mutant in RT (ΔRT), which resulted in translation of <100 aa of RT, were used as negative controls. Wild-type proviral DNA served as the positive control. Viral proteins from transfected cells were immunoblotted with anti-CA antibody (Fig. 5B). All mutants produced viral proteins. MA-1391, p12-1518, p12-1542, p12-1587, and CA-1848 were similar to wild type. p12-1542 had greater amounts of unprocessed Gag and CA-1794 had more unprocessed Pr65gag relative to CA in the cells. Virions harvested from these cells were also immunoblotted (Fig. 5C). All mutants produced virions. Mutants MA-1391, p12-1542, and p12-1587 processed their Gag proteins similar to wild type. Mutants CA-1794 and CA-1848 had significant amounts of processing intermediates in the virion. A qualitatively similar processing pattern, though to a lesser extent, was seen with p12-1518. However, all mutants except p12-1542 were found to have RT activity similar to wild type (Fig. 5D), indicating that virions were assembled, released, and contained RT. p12-1542 has somewhat lower (26%) RT activity. As expected, Δgag produced neither full-length Gag nor virions, and ΔRT, while producing appropriately processed Gag proteins of correct sizes, did not score positive for RT activity.
Fig. 5.
Stepwise conventional analysis of a few mutations. (A) MoMLV life cycle, focusing on specific steps that were analyzed for each mutant. (B–I) Data from each mutant are displayed in vertical rows, with each subsequent step represented directly below the previous one. Wild-type (WT) virus, gag deletion mutant (Δgag), and RT deletion mutant (ΔRT virus) were used as controls. Mock controls underwent identical treatments, except no proviral DNA was used in the transfection. (B) Expression of viral proteins in transfected cells. Cell lysates from transfected cells were analyzed by Western blots using anti-CA antibody. Arrows indicate positions of Gag precursor Pr65gag and CA. (C) Virion proteins. Virions harvested from transfected cells were analyzed by Western blots as in B.(D) Viral assembly and particle release. Cell supernatants were collected 48 and 72 h posttransfection and analyzed for RT activity. (E) PCR amplification of minus-strand strong-stop DNA [(–)ss DNA]. Primers amplify a 124-bp stretch of the R-U5 region. (F) PCR amplification of gag DNA, using one primer within the insertion and a second in the CA region. For WT virus and for plasmid pNCA, which do not contain insertions, no PCR product was seen. (G) PCR amplification of the LTR junction. Arrows indicate the position of the LTR-junction product. For the pNCA plasmid, primers amplify the β-lactamase gene located in the region between the two LTRs (arrowhead). (H) Amplification of mtDNA. (I) Viral infectivity measured by RT activity. Culture supernatants from infected Rat2 cells were harvested at day 4 and day 6 postinfection and assayed for RT activity.
Virions from these transfected cells were used to infect Rat2 cells. Low molecular weight DNA from Rat2 cells was used as a template for PCR designed to detect viral-replication intermediates. Heat-inactivated virus was used as a control in infection to confirm that PCR products were a result of amplification of DNA produced by virally mediated reverse transcription and not due to contaminating plasmid DNA carried over from the transfections (not shown). Parent proviral plasmid (pNCA) was used as another control. Mutants MA-1391, p12-1542, and p12-1587 were capable of reverse transcription as indicated by a PCR designed to detect the region included in the minus-strand strong-stop DNA (Fig. 5E). This finding was confirmed by a second PCR that amplified the gag region of viral DNA by using a primer within gag and a second primer within the insertion (Fig. 5F). The size of this PCR product varied with the location of the insertion. Because neither the wild-type virus nor pNCA contained an insertion, they did not generate a PCR product in this amplification. The CA mutants and p12-1518 did not appear to perform reverse transcription, suggesting that they might be impaired in viral entry or uncoating. To assay nuclear transport, we amplified the LTR junction region (Fig. 5G), because the presence of circular viral DNA containing the LTR junction is a hallmark of retroviral DNA transport into the nucleus (20, 21). Of the three mutants that could perform reverse transcription, only MA-1391 and p12-1587 were able to enter the nucleus. Mutant p12-1542, although able to perform reverse transcription, was most likely unable to enter the nucleus, suggesting that an insertion at nucleotide 1542 disrupts a domain of p12 essential for nuclear import of viral DNA. Previous studies have shown mutants of p12 in nucleotides 1536–1552 (and also in nucleotides 1505–1517) to be defective in nuclear transport (22). PCR amplification using rat mtDNA primers showed that approximately equal amounts of DNA were used as templates in the various PCRs (Fig. 5H). Mutants MA-1391 and p12-1587, which were similar to wild type for each tested function, also resulted in wild-type levels of RT activity in the virions released from infected cells (Fig. 5I), suggesting that insertions at these locations did not disrupt viral function. In summary, our detailed analysis of selected mutants permitted us to assign specific roles to the corresponding positions in gag sequence.
Discussion
Our results with genetic footprinting agree with previous data on Gag structure and function. We have, in addition, identified regions that play an essential role in viral replication. Regions at the border of MA/p12 and at p12/CA were found to be essential for replication, suggesting that the proteolytic cleavage sites are important. Some regions of p12 that have been shown to play a role in viral release and others in transport of viral DNA into the nucleus lay in or close to a footprint in p12. The N terminus of MoMLV CA has not been a focus of mutagenesis before this study. We find that the entire region is unable to accept insertions and is thus essential for replication. The function of other novel small footprints in MA and p12 is not yet understood. An apparent discrepancy between previously published mutagenesis and our footprinting results involves serine residues at 1643 and 1694 in p12. These serine residues are known to play a role in phosphorylation of p12, and when changed to alanine, result in severely impaired virions (15). These regions appear to tolerate insertions well in our study, perhaps because unlike in alanine-substitution mutagenesis, insertions do not remove the serine residues and may permit phosphorylation.
Our mutagenesis revealed several regions within gag where the density of insertions generated by integrase was high, such as the C-terminal third of MA or portions of p12. This finding is somewhat related to the enzyme used for integration or transposition, enzymatic reaction conditions, and the specific characteristics of the sequence as it interacts with the enzyme (23). As far as we can predict, the density of generated mutations in a given sequence is unrelated to the role of that sequence in infection.
We found that large regions of gag appear to tolerate 36-nt insertions. Although one might intuitively expect retroviral genes to have evolved to be small, efficient, and intolerant of further changes, that is not what we observed. Most of the C terminus of MA and large portions of p12 could tolerate 36-nt insertions, suggesting that large contiguous features in this region that are essential for replication are unlikely. This finding also suggests suitable places to insert epitope tags that could be used for isolating or localizing replication intermediates. Because these locations accept 12-aa insertions, they are likely to accept other small tags such as the FLAG tag (8 aa) or the (His)n tag. Some might even accept a larger tag, such as GFP. Genetic footprinting using epitope tags as insertions would be an efficient method to generate replication-competent, tagged virus. Such a virus would be useful for intracellular localization and purification of replication intermediates.
We saw a high degree of variation in the sizes of footprints in different regions of the viral sequence. With p12 and MA, footprints extended to a few nucleotides, consistent with studies that have found p12 and MA to be tolerant of mutations (2). In contrast, the entire N-terminal half of CA was intolerant of 12-aa insertions. Little is known about this region from previous mutagenesis. However, a linker-insertion mutagenesis that selected for replication-competent virus found 3 of the 28 viable mutations in p12, and none in CA (16). Some of the difference between tolerance to insertions seen with MA or p12 and that with CA might be attributed to changing Gag–Gag interactions as the capsids mature by proteolytic cleavage. MA–MA contacts and NC–NC contacts appear to remain the same on maturation, but CA–CA contacts change (24, 25). Thus, it is possible that there are greater constraints on CA structure, making it less likely to be tolerant of insertions. Smaller insertions or substitution mutations might be better tolerated in CA. Also, with CA possibly involved in many steps of the viral life cycle, genetic footprinting will likely provide most information when selections for specific stages of viral replication are performed.
Any selection where recovery of a DNA or RNA molecule depends on a property of the sequence or its encoded products is amenable to genetic footprinting. For example, the selected MoMLV library could consist of viral nucleic acid that is resistant to RNase, providing a method to select for mutants capable of viral DNA synthesis. Similarly, isolation of circular DNA would provide a means of selecting mutants capable of nuclear transport. Footprinting with such biochemically selected material would allow the functional mapping of viral sequences for specific stages in the viral life cycle. This approach can be used to analyze an entire viral genome to further elucidate the roles of viral sequences in specific steps of replication. For many viruses, a map that defines essential regions in the genome would be very useful to understand the roles of many components of viral replication, and also to generate viruses defective in multiple essential regions, for making vaccines or for identifying target sites for small-molecule inhibitors of replication.
Acknowledgments
I.R.S. thanks Pat Brown for his support at the inception and early stages of this project. We are grateful to Steve Goff for numerous insightful discussions and to Eran Bacharach, G. Gao, and Andrew Yueh for technical advice. We thank Denise De Las Neuces for screening CA mutants. We thank Harsh Thaker and Brett Lauring for valuable comments on the manuscript. This work was supported by National Institutes of Health Grant K08-AI01678 (to I.R.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RT, reverse transcriptase; moi, multiplicity of infection; MoMLV, Moloney murine leukemia virus; CA, capsid; MA, matrix; NC, nucleocapsid.
References
- 1.Leis, J., Baltimore, D., Bishop, J. M., Coffin, J., Fleissner, E., Goff, S. P., Oroszlan, S., Robinson, H., Skalka, A. M., Temin, H. M., et al. (1988) J. Virol. 62, 1808–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swanstrom, R. & Wills, J. W. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 263–334.
- 3.Singh, I., Crowley, R. & Brown, P. (1997) Proc. Natl. Acad. Sci. USA 94, 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinonez, R., Sinha, I., Singh, I. R. & Sutton, R. E. (2003) Virology 307, 98–115. [DOI] [PubMed] [Google Scholar]
- 5.Laurent, L. C., Olsen, M. N., Crowley, R. A., Savilahti, H. & Brown, P. O. (2000) J. Virol. 74, 2760–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothenberg, S. M., Olsen, M. N., Laurent, L. C., Crowley, R. A. & Brown, P. O. (2001) J. Virol. 75, 11851–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colicelli, J. & Goff, S. P. (1988) J. Mol. Biol. 199, 47–59. [DOI] [PubMed] [Google Scholar]
- 8.Telesnitsky, A., Blain, S. & Goff, S. (1995) Methods Enzymol. 262, 347–362. [DOI] [PubMed] [Google Scholar]
- 9.Lim, D., Orlova, M. & Goff, S. P. (2002) J. Virol. 76, 8360–8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirt, B. (1967) J. Mol. Biol. 26, 365–369. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, F. & Hallet, B. (2000) Trends Microbiol. 8, 571–577. [DOI] [PubMed] [Google Scholar]
- 12.Gonsky, J., Bacharach, E. & Goff, S. P. (2001) J. Virol. 75, 2616–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan, B., Fassati, A., Yueh, A. & Goff, S. P. (2002) J. Virol. 76, 10801–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan, B., Campbell, S., Bacharach, E., Rein, A. & Goff, S. P. (2000) J. Virol. 74, 7250–7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yueh, A. & Goff, S. P. (2003) J. Virol. 77, 1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobel, L. I. & Goff, S. P. (1984) Proc. Natl. Acad. Sci. USA 81, 4149–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu, H. W., Schwartzberg, P. & Goff, S. P. (1985) Virology 142, 211–214. [DOI] [PubMed] [Google Scholar]
- 18.Alin, K. & Goff, S. P. (1996) Virology 222, 339–351. [DOI] [PubMed] [Google Scholar]
- 19.Wang, M. Q. & Goff, S. P. (2003) J. Virol. 77, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis, J. & Bernstein, A. (1989) J. Virol. 63, 2844–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobel, L. I., Murphy, J. E. & Goff, S. P. (1989) J. Virol. 63, 2629–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan, B., Li, X. & Goff, S. P. (1999) EMBO J. 18, 4700–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu, R. & Grandgenett, D. P. (2003) J. Virol. 77, 6482–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pepinsky, R. (1983) J. Biol. Chem. 258, 11229–11235. [PubMed] [Google Scholar]
- 25.Pepinsky, R., Cappiello, D., Wilkowski, C. & Vogt, V. (1980) Virology 102, 205–210. [DOI] [PubMed] [Google Scholar]