Abstract
The ability of a pathogen to cause infection depends on successful colonization of the host, which, in turn, requires adaptation to various challenges presented by that host. For example, host immune cells use a variety of mechanisms to control infection by bacterial pathogens, including the production of bactericidal reactive oxygen species. Prokaryotic and eukaryotic cells have developed ways of protecting themselves against this oxidative damage; for instance, Borrelia burgdorferi alters the expression of oxidative-stress-related proteins, such as a Dps/Dpr homolog NapA (BB0690), in response to increasing levels of oxygen and reactive oxygen species. These stress-related genes appear to be regulated by a putative metal-dependent DNA-binding protein (BB0647) that has 50.7% similarity to the peroxide-specific stress response repressor of Bacillus subtilis, PerR. We overexpressed and purified this protein from Escherichia coli and designated it Borrelia oxidative stress regulator, BosR. BosR bound to a 50-nt region 180 bp upstream of the napA transcriptional start site and required DTT and Zn2+ for optimal binding. Unlike the Bacillus subtilis PerR repressor, BosR did not require Fe2+ and Mn2+ for binding, and oxidizing agents, such as t-butyl peroxide, enhanced, not eliminated, BosR binding to the napA promoter region. Surprisingly, transcriptional fusion analysis indicated that BosR exerted a positive regulatory effect on napA that is inducible with t-butyl peroxide. On the basis of these data, we propose that, despite the similarity to PerR, BosR functions primarily as a transcriptional activator, not a repressor of oxidative stress response, in B. burgdorferi.
During its infectious cycle Borrelia burgdorferi, the causative agent of Lyme disease, survives and proliferates in both an arthropod and a mammalian host. One of the many challenges B. burgdorferi faces during this infectious cycle is from reactive oxygen and nitrogen species generated by the host immune cells. These reactive oxygen species (ROS), such as superoxide radicals ( ), hydrogen peroxide (H2O2), and hydroxyl radicals (•OH), and reactive nitrogen species, such as nitric oxide (NO) and peroxynitrite, can damage DNA, RNA, proteins, and lipids (1) and are deleterious to bacterial cells. To protect against this damage, bacteria express proteins [e.g., superoxide dismutase (SOD), alkylhydroperoxide reductase (AhpR), catalase/peroxidases, etc.] that detoxify the reactive species and repair the damage caused by them.
), hydrogen peroxide (H2O2), and hydroxyl radicals (•OH), and reactive nitrogen species, such as nitric oxide (NO) and peroxynitrite, can damage DNA, RNA, proteins, and lipids (1) and are deleterious to bacterial cells. To protect against this damage, bacteria express proteins [e.g., superoxide dismutase (SOD), alkylhydroperoxide reductase (AhpR), catalase/peroxidases, etc.] that detoxify the reactive species and repair the damage caused by them.
Bacteria adapt to the presence of ROS by increasing the expression of detoxification enzymes. These responses are coordinated by transcription factors that regulate target genes in response to ROS. The best characterized systems in bacteria are the OxyR and SoxR/SoxS oxidative stress response systems of Escherichia coli, which activate antioxidant genes in response to H2O2 and to  , respectively, and the peroxide-sensitive repressor PerR from Bacillus subtilis. The OxyR protein is directly sensitive to oxidation, forming an intramolecular disulfide bond upon oxidation, and only this oxidized form is capable of activating transcription of genes in the oxidative stress response (2). Positive regulation of genes involved in the glutaredoxin system results in the reduction of OxyR, returning the regulator to an inactive, thiol-containing form (3). The SoxR protein contains a redox-active Fe–S cluster that regulates DNA binding. Once oxidized, SoxR induces transcription of soxS and SoxS activates transcription of genes in the SoxR/S regulon (4). The PerR protein is a metalloprotein that binds one or more copies of a conserved, 14-bp inverted repeat to repress expression of target genes (5). The oxidation of the Zn2+,Fe2+ form of PerR (active) by H2O2 abolishes binding, causing derepression of target genes.
, respectively, and the peroxide-sensitive repressor PerR from Bacillus subtilis. The OxyR protein is directly sensitive to oxidation, forming an intramolecular disulfide bond upon oxidation, and only this oxidized form is capable of activating transcription of genes in the oxidative stress response (2). Positive regulation of genes involved in the glutaredoxin system results in the reduction of OxyR, returning the regulator to an inactive, thiol-containing form (3). The SoxR protein contains a redox-active Fe–S cluster that regulates DNA binding. Once oxidized, SoxR induces transcription of soxS and SoxS activates transcription of genes in the SoxR/S regulon (4). The PerR protein is a metalloprotein that binds one or more copies of a conserved, 14-bp inverted repeat to repress expression of target genes (5). The oxidation of the Zn2+,Fe2+ form of PerR (active) by H2O2 abolishes binding, causing derepression of target genes.
Analysis of the genome has revealed that B. burgdorferi contains genes encoding a Mn-dependent superoxide dismutase (sodA) and a neutrophil activating protein, BB0690 (napA), a Dps (DNA-binding protein from starved cells)/Dpr (Dps-like peroxide resistance gene) homolog (www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gbb) that complements an AhpR mutant of E. coli. However, the regulatory proteins controlling these genes have not been characterized. An ORF that has 50.7% similarity to a protein encoded by perR from Bacillus subtilis has been identified (see the web site given above). PerR, the central regulator of peroxide-specific stress response in Bacillus, has been shown to regulate genes that are involved in oxidative stress response, such as those encoding AhpR (6) and the major vegetative catalase (katA) (7). Based on the similarity between the B. burgdorferi ORF BB0647 and Bacillus subtilis PerR, we hypothesized that these proteins have similar functions and began to characterize the former protein. Our working hypothesis is that B. burgdorferi cells are using the two proteins SOD and NapA to convert  to H2O2 and then to H2O in a two-step process (unpublished results) that is regulated by BB0647, designated BosR.
to H2O2 and then to H2O in a two-step process (unpublished results) that is regulated by BB0647, designated BosR.
The purification of BosR and studies of its effect on gene expression are reported here. Mobility-shift DNA-binding assays indicated that BosR requires Zn2+ for maximal binding and can bind DNA in both an oxidized and a reduced state. Transcriptional fusion data indicated that BosR exerts a positive regulatory effect on napA transcription that is inducible with t-butyl peroxide. Surprisingly, despite the significant similarity of BosR to an oxidative stress repressor, our data suggest that BosR functions as an activator of napA transcription, and it represents a class of oxidative stress regulators not previously recognized.
Materials and Methods
Bacterial Strains. Low-passage B. burgdorferi strains B31 and B31 A3 (8) were grown in modified Barbour–Stoenner–Kelly (BSK-II) medium (9) at 34°C under an atmosphere of 4% H2/5% CO2/91% N2, and cell numbers in each culture were determined by dark-field microscopy. E. coli Rosetta strain [BL21 (DE3)/(pLysS)] (Invitrogen) cells were grown in LB medium or M9 minimal medium (10) at 37°C. Ampicillin (50 μg/ml), chloramphenicol (30 μg/ml), or isopropyl β-d-thiogalactopyranoside (IPTG) (1.0 mM) was added as needed. All chemicals were purchased from Sigma, and DNA restriction or modifying enzymes were purchased from Promega unless otherwise stated.
Overexpression and Purification of BosR. The bosR ORF was amplified by PCR from B. burgdorferi strain B31 by using the forward primer 5′-GTGGGATCCATGAACGACAACAT-3′ and reverse primer 5′-GTGGGATCCCCTCGGACTAACCAGT-3′, generating a BamHI site at each end of the PCR product (underlined). This fragment was digested with BamHI and cloned into the expression vector pET-3a (Novagen), generating pJAB3. pJAB3 was introduced into E. coli Rosetta strain [BL21 (DE3)/(pLysS)] (Invitrogen) with selection for resistance to chloramphenicol (35 μg/ml) and ampicillin (50 μg/ml).
For overexpression, cells containing pJAB3 were grown in 500 ml of LB/1% glucose at 37°C with shaking. When cells reached an OD600 of 0.6, expression of BosR was induced with 1 mM IPTG for 4 h. Cells were harvested by centrifugation (8,000 × g, 15 min, room temperature), resuspended in lysis buffer (20 mM Tris·HCl, pH 7.0/5% glycerol/1 mM DTT), and lysed by three passages through a cold French press cell at 14,000 psi (96 MPa). The inclusion bodies were harvested by centrifugation (12,000 × g, 30 min, 4°C), suspended in lysis buffer containing 2% deoxycholate, incubated with gentle agitation for 60 min at 4°C, and harvested (30,000 × g, 30 min, 4°C). The pellet containing the inclusion bodies was resuspended in 25 ml of lysis buffer that contained 0.25% Sarkosyl and was incubated for 1 h at 4°C with gentle agitation. The insoluble material was removed by centrifugation (12,000 × g, 30 min, 4°C), and the supernatant was dialyzed against 4 liters of lysis buffer for 12 h at 4°C. The extract was applied to a HiTrap Heparin HP (Amersham Pharmacia) column equilibrated with lysis buffer at 4°C, and proteins were eluted by using a linear 0–1.0 M NaCl gradient. One-milliliter fractions containing BosR, as determined by SDS/PAGE, were pooled and concentrated by using Centriplus YM-10 (Millipore). Concentrated protein was desalted by using a PD-10 desalting column (Amersham Pharmacia), applied to a HiTrap Q anion-exchange column (Amersham Pharmacia) at 4°C, and eluted by using a linear gradient of 0–1.0 M NaCl, and the protein was stored in 5% glycerol at –80°C. Purified BosR was used to raise polyclonal antiserum in a female New Zealand White rabbit at Cocalico (Reamstown, PA). The sequence was determined by N-terminal amino acid analysis by the Molecular Genetics Instrumentation Facility, University of Georgia (Athens).
Mobility-Shift DNA-Binding and DNase I Footprinting. The napA promoter/operator (P/O) region was amplified by PCR from B. burgdorferi B31 by using the forward primer 5′-GTGGAATTCTTTGCATTGGGGTTGTG-3′ and the reverse primer 5′-GTGGAAT TCTTGCGTCTAAATCATCCTT-3′, generating a 380-bp fragment. The fragment was gel-purified and end-labeled by using T4 polynucleotide kinase and [γ-32P]ATP. Binding reactions were carried out in a 20-μl volume that contained binding buffer (20 mM Tris·HCl, pH 7.5/5% glycerol/1 mM DTT/50 μg/ml BSA/50 μg/ml salmon sperm DNA/1 mM MgCl2), purified BosR, and 20,000 cpm of target sequence (≈10–20 fmol). The reactions were incubated for 10 min at room temperature, and the products were separated on a nondenaturing 6% polyacrylamide gel (25 mA constant amps, 2 h) and analyzed by autoradiography. To determine metal selectivity of BosR–DNA interaction, binding reactions were performed as described above with the addition of 10 μM indicated divalent metal ion. Stock solutions of metals (MnCl2, ZnCl2, or FeSO4) were made fresh before each assay. The effects of DTT and t-butyl peroxide on BosR–DNA interaction were determined by mobility-shift DNA-binding assays. Binding reactions were performed as above in the absence of DTT, in the presence of 100 mM DTT, or in the presence of 1 mM DTT and 1 mM t-butyl peroxide. All reactions were performed with 10 μM ZnCl2.
DNase I protection assays were performed essentially as described (11). The napA P/O region, extending from –200 to +200, was amplified from B. burgdorferi B31 chromosomal DNA by using primers NapAP5 (5′-TTCAAGGGTATCATCTATTTCTAG-3′) and NapAP3 (5′-GTGAATAACAAAGAAATTGGTATC-3′) with Taq DNA polymerase. The PCR product was cloned into pCR2.1 (Invitrogen) by the T/A cloning procedure to generate pJP12 and digested with XbaI. The linearized plasmid JP12 was end-labeled by using T4 polynucleotide kinase and [γ-32P]ATP and digested with BamHI, releasing a 490-bp fragment containing the P/O region of napA. The napA P/O region was isolated from a nondenaturing 5% polyacrylamide gel. A 20-μl reaction mixture containing 20 mM Tris·HCl (pH 7.6), 40 mM KCl, 5 mM MgCl2, 1 mM CaCl2, 1 mM DTT, 5% glycerol, 1 μg BSA, target sequence DNA (20,000 cpm), and purified BosR was incubated at 30°C for 15 min. The mixture was then treated with DNase I (0.025 unit) at 30°C for 1 min, and the reaction was stopped by the addition of 20 μl of stop buffer (5 M ammonium acetate/50 mM EDTA/50 mg/ml tRNA). The DNA was precipitated with ethanol, separated on a denaturing 6% polyacrylamide gel, and analyzed by autoradiography.
β-Galactosidase Assays. To generate the napAP/O-lacZ transcriptional fusion, the 4.7-kb BamHI fragment containing a promoterless lacZ and aph (kanamycin-resistance marker) from pKOK6 was cloned into the BamHI site of pJP12 to generate pJP13. The transcriptional fusion was released by digestion of pJP13 with KpnI and treated with T4 DNA polymerase to create blunt ends. The fragment was digested with XbaI and ligated into the XbaI and EcoRV sites of pACYC184 to generate pnapAP/O-lacZ.All constructs were confirmed by sequencing.
The β-galactosidase activity from cells harboring transcriptional fusions was measured as described by Miller (12). E. coli cells harboring pnapAP/O-lacZ fusion coresident with pJAB3 were grown in LB, LB plus IPTG, M9 minimal medium, or M9 plus IPTG at 37°C to mid-logarithmic phase (OD600 ≈ 0.4–0.5). Cells were harvested by centrifugation (5,000 × g, 5 min) and assayed for enzyme activity. To test the effect of peroxides on expression, cells were grown to mid-logarithmic phase and induced with IPTG for 1 h, the cultures were split, and half the cells were exposed to 50 μM H2O2 or 100 μM t-butyl peroxide for 30 min. After the incubation, the cells were assayed for enzyme activity.
NapA Induction with t-Butyl Peroxide. A 1-liter culture of B. burgdorferi strain B31 A3 (8) was grown to a density of 5 × 107 cells per ml under an atmosphere of 4% H2/5% CO2/91% N2 and split into five 200-ml cultures. Each 200-ml culture was treated with t-butyl peroxide at concentrations ranging from 0 to 1 mM for 8 h. After exposure, cells were harvested by centrifugation (5,000 × g, 15 min), washed with Hepes buffer (20 mM Hepes/50 mM NaCl, pH 7.6), and resuspended in 2× SDS loading buffer (100 mM Tris·HCl, pH 6.8/4% SDS/0.2% bromophenol blue/20% glycerol/200 mM 2-mercaptoethanol).
Electrophoresis and Immunoblotting. Proteins were separated on SDS/12% polyacrylamide gels. For immunoblotting, proteins were electrophoretically transferred to nitrocellulose (0.2-μm pore size TransBlot transfer membrane; Bio-Rad). Membranes were blocked with 5% nonfat dry milk in PBS solution overnight at 4°C (13). The primary antibody, α-NapA (generated from purified NapA protein by Cocalico) or α-BosR, was applied to the blot and incubated 1 h at room temperature. The blot was then washed three times in TTBS (100 mM Tris·HCl, pH 7.5/150 mM NaCl/0.1% Tween 20) for 10 min each to remove primary antibody. Secondary antibody (alkaline phosphatase-conjugated goat anti-rabbit IgG) was diluted 1:5000 in TTBS and applied to the blot for 1 h at room temperature, then washed three times with TTBS. The blot was visualized by using chemiluminesence (ECL Plus Western Blotting Detection Reagents, Amersham Pharmacia).
Results
Purification of BosR. Preliminary analysis of protein profiles from B. burgdorferi cell extracts indicated that exposure to increasing concentrations of O2, H2O2, or t-butyl peroxide affected protein levels both positively and negatively (data not shown). This observation suggested that a regulatory effector (or effectors) was altering gene expression in response to oxygen concentration and/or to negate damage by ROS. Analysis of the B. burgdorferi genome sequence revealed a potential ROS-dependent regulatory protein. ORF BB0647 has 50.7% similarity to PerR, a peroxide stress response repressor from Bacillus subtilis (www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gbb). To assess its potential role in the oxidative stress response of B. burgdorferi, BB0647 [designated Borrelia oxidative stress regulator (bosR)] was cloned and the protein was overexpressed in E. coli, producing a 22-kDa protein that localized to inclusion bodies in the cell lysate. BosR from inclusion bodies was solubilized with 0.25% Sarkosyl and purified with a heparin-agarose affinity column followed by ion-exchange chromatography. This procedure yielded BosR that was purified to apparent homogeneity as determined by SDS/PAGE (data not shown). N-terminal amino acid analysis of the purified protein indicated that the correct protein had been purified.
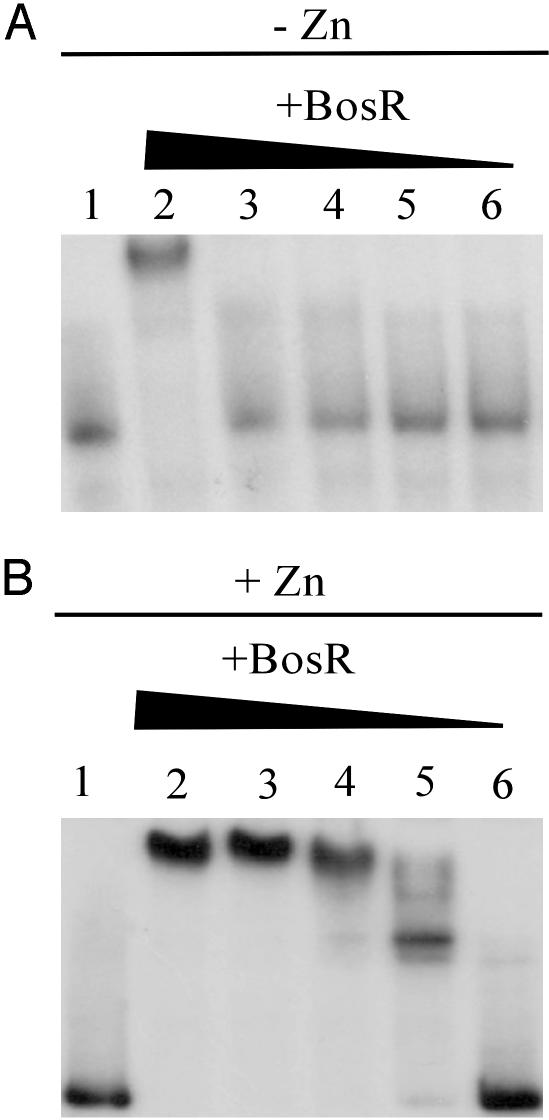
BosR Bound the napA P/O Region. Two lines of evidence suggested that NapA expression could be affected by increased O2 or ROS and regulated by BosR. First, NapA is homologous to the Dps/Dpr family of proteins and complements an AhpR mutant of E. coli (unpublished results). Both Dps/Dpr and AhpR are commonly regulated by ROS-dependent repressors and activators in bacteria (14). Second, a sequence immediately upstream of napA (15 bp upstream of the actual translation start site) is similar to the consensus binding sequence for PerR from Bacillus subtilis (5). To test the hypothesis that BosR regulates napA expression, purified BosR was assayed for the ability to bind the napA P/O by using a mobility-shift DNA-binding assay. BosR bound to the probe at concentrations of 1 μM or higher in the absence of divalent metals (Fig. 1A, lane 2). However, in the presence of 10 μM Zn2+, greater than 90% of the probe was shifted with 300 nM protein (Fig. 1B, lane 5). Other divalent metal ions such as Fe2+ and Mn2+ (final concentrations of 10 μM) did not enhance the binding of BosR to the target sequence. The addition of Mn2+ to reaction mixtures containing BosR and Zn2+ abolished binding (data not shown), whereas the addition of Fe2+ had no effect. These data indicate that Zn2+ alone was required for efficient binding of BosR to the target sequence.
Fig. 1.
BosR bound to a DNA fragment that contained the napA P/O region, as shown by autoradiographs of mobility-shift DNA-binding assays using purified BosR and 32P-labeled napA P/O region. Decreasing concentrations of BosR (1,000, 700, 500, 300, and 100 nM; lanes 2–6, respectively) were incubated with labeled probe and 1 mM DTT in the absence of Zn2+ (A) or in the presence of 10 μM Zn2+ (B). Lane 1 in both A and B contained no BosR.
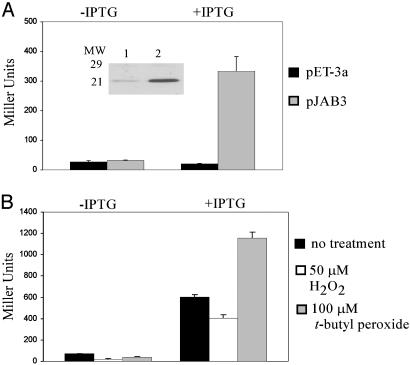
BosR Activates Transcription of napAP/O-lacZ in E. coli. To determine whether BosR was negatively or positively affecting transcription of napA,a napAP/O-lacZ transcriptional fusion was constructed and introduced into E. coli strain BL21 (DE3)/(pLysS) harboring pJAB3, and β-galactosidase activity was measured under various conditions (Fig. 2). In cells harboring pnapAP/O-lacZ and the control vector pET-3a, <30 units of β-galactosidase activity was detected (Fig. 2 A, black bar). Without induction of BosR in cells harboring pnapAP/O-lacZ and pJAB3, β-galactosidase activity was equivalent to activity measured in cells harboring the control vector (Fig. 2 A, –IPTG, gray bar). When BosR was induced in these cells, β-galactosidase activity increased (Fig. 2 A, +ITPG, gray bar), indicating that BosR exerted a positive regulatory effect on the napAP/O-lacZ transcriptional fusion. Immunoblot analysis indicated that BosR is expressed at much higher levels in cells when induced with IPTG (Fig. 2 A Inset, lane 2), compared with uninduced cells (Fig. 2 A Inset, lane 1). Interestingly, the transcriptional fusion data suggested that BosR acts as an activator of napA expression rather than a repressor.
Fig. 2.
BosR activates transcription of a napAP/O-lacZ fusion in vivo. (A) β-Galactosidase activity was measured in E. coli cells harboring pnapAP/O-lacZ fusion and pET-3a (black bar) or pJAB3 (gray bar). IPTG was added to induce the expression of BosR. (Inset) Immunoblot analysis of E. coli cells harboring pnapAP/O-lacZ fusion and pJAB3 grown in M9 (lane 1) and M9 + IPTG (lane 2) probed with α-BosR antibody. Molecular weight markers (× 10–3) are indicated in the margin. (B) E. coli cells harboring pnapAP/O-lacZ fusion and pJAB3 (black bar) were grown to mid-logarithmic phase in M9 or M9 + IPTG and exposed to 50 μMH2O2 (white bar) or 100 μM t-butyl peroxide (gray bar) and assayed for β-galactosidase activity.
The expression of most oxidative stress genes in bacteria increases dramatically in response to elevated levels of peroxides or other oxidants. To test the effects of peroxides on the interaction of BosR and napA P/O, the reporter constructs described above were challenged with hydrogen peroxide or t-butyl peroxide, and β-galactosidase activity was measured (Fig. 2B). The cells were grown to OD600 of ≈0.5, BosR was induced with IPTG for 1 h, and then cells were exposed to various peroxides for 30 min. In uninduced samples, β-galactosidase activity was <75 units (Fig. 2B, –IPTG), whereas the activity increased to >600 units after the induction of BosR (Fig. 2B, +IPTG, black bar). Interestingly, β-galactosidase activity increased 2-fold upon addition of t-butyl peroxide (Fig. 2B, +IPTG, gray bar). Surprisingly, no effect on β-galactosidase activity was seen upon addition of hydrogen peroxide (Fig. 2B, +IPTG, white bar). Because these cells were grown with aeration to maximize BosR induction, intracellular concentrations of hydrogen peroxide, resulting from the incomplete reduction of oxygen in aerobic metabolism (15), may have reached levels that affected initial BosR activity. These data indicated BosR interactions with the napAP/O-lacZ was enhanced by ROS and maximal activity was obtained after exposure to t-butyl peroxide.
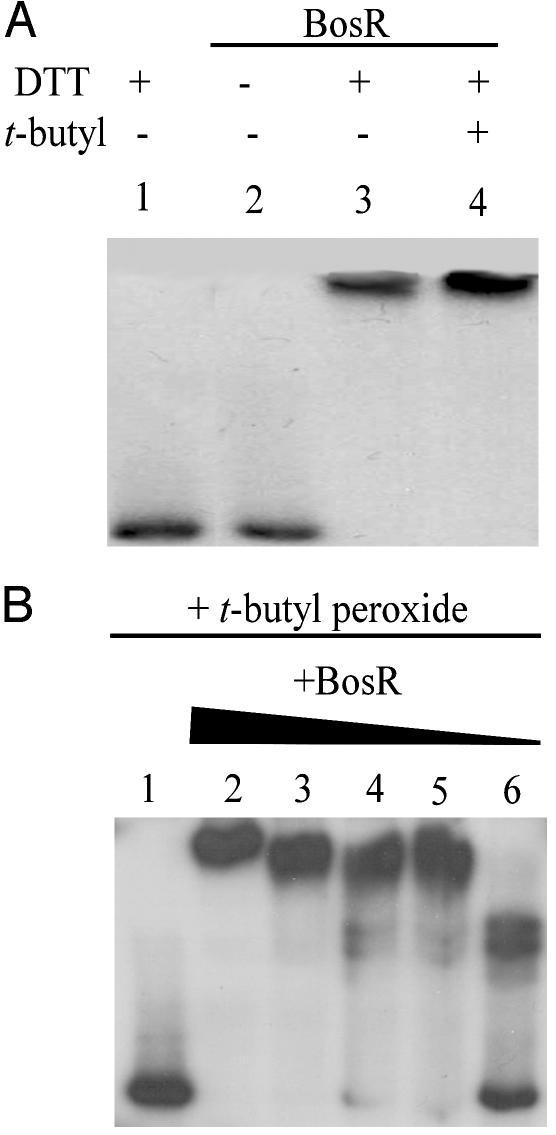
BosR Bound napA P/O in Both Oxidized and Reduced States. To determine the effects of oxidizing agents on BosR binding to target DNA, labeled probe was incubated with BosR in the absence of DTT (Fig. 3A, lane 2), in the presence of 100 mM DTT (Fig. 3A, lane 3) or in the presence of 1 mM DTT and 1 mM t-butyl peroxide (Fig. 3A, lane 4). In the presence of 100 mM DTT, BosR bound the napA promoter, indicating that BosR was able to bind DNA in a reduced state (Fig. 3A, lane 3). When no DTT was added, no binding was observed (Fig. 3A, lane 2). Moreover, when BosR–DNA complexes were challenged with 1 mM t-butyl peroxide, binding was completely unaffected (Fig. 3A, lane 4). Interestingly, binding of BosR could be detected at concentrations as low as 100 nM (Fig. 3B, lane 6) when decreasing amounts of BosR were challenged with 1 mM (or as high as 10 mM) t-butyl peroxide. However, in the absence of t-butyl peroxide, binding could be detected only at concentrations ≥300 nM. These data suggest that BosR required DTT for initial binding to the napA P/O and that binding was enhanced, not eliminated, by the addition of 10× excess of an oxidizing agent (t-butyl peroxide).
Fig. 3.
BosR bound napA P/O in both oxidized and reduced states, as shown by autoradiographs of mobility-shift DNA-binding assays using purified BosR and 32P-labeled napA P/O region. In A, 700 nM BosR was incubated with labeled probe in the absence of DTT (lane 2), in the presence of 100 mM DTT (lane 3), or in the presence of 1 mM DTT and 1 mM t-butyl peroxide (lane 4). Lane 1 contained no BosR. In B, decreasing concentrations of BosR (1,000, 700, 500, 300, and 100 nM; lanes 2–6) were incubated with labeled probe and 1 mM DTT in the presence of 1 mM t-butyl peroxide. Lane 1 contained no BosR.
t-Butyl Peroxide Induces Expression of napA in Vivo. Using E. coli β-galactosidase reporter assays, we have demonstrated that BosR activates expression of napA, and this expression is inducible with t-butyl peroxide. To correlate these data with the expression of napA in B. burgdorferi, B. burgdorferi strain B31 A3 cells were grown anaerobically under an atmosphere of 4% H2/5% CO2/91% N2 to a cell density of 5 × 107 cells per ml. The culture was divided and treated with various concentrations of t-butyl peroxide (0, 0.25, 0.5, 0.75, and 1 mM) for 8 h at 34°C. Equivalent amounts of protein from each aliquot of cells was separated by SDS/PAGE, transferred to nitrocellulose, and probed with either α-NapA or α-BosR antiserum (Fig. 4). An increase in NapA was observed when cells were treated with 0.5 mM t-butyl peroxide (Fig. 4A, lane 4) compared with untreated cells (Fig. 4A, lane 2), and NapA levels increased as t-butyl peroxide concentration increased (Fig. 4A, lanes 3–6). Interestingly, there appeared to be no significant change in the levels of BosR in the same samples (Fig. 4B, lanes 2–6). This observation suggested that bosR is not up-regulated in response to t-butyl peroxide and that the increase in NapA was due not to an increase in BosR but to an increase in the amount of active BosR.
Fig. 4.
t-Butyl peroxide induced expression of napA in vivo. Shown are immunoblots of B. burgdorferi B31 A3 cells grown in an atmosphere of 4% H2/5% CO2/91% N2 and treated with increasing concentrations of t-butyl peroxide (0.25, 0.5, 0.75, and 1 mM; lanes 3–6) for 8 h. Proteins were separated by SDS/PAGE, transferred to nitrocellulose, and probed with rabbit α-NapA antibody (A) or rabbit α-BosR antibody (B). Lane 2 contained untreated B31 A3 cells. Lane 1 contained purified NapA (A) or purified BosR (B). Molecular weight (×10–3) markers are indicated on the left.
BosR Bound to a 50-bp Sequence 190 bp Upstream of the napA Transcriptional Start Site. Mobility-shift assays indicated that in the presence of 10 μM Zn2+, BosR bound to a DNA fragment containing the napA P/O. To determine the sequence recognized by BosR within this P/O region, DNase I footprint analysis was performed. A 490-bp DNA fragment excised from pJP12 containing the –200 to +200 region of napA was labeled on the coding strand. BosR protected a 50-nt region on the coding strand from –137 to –187 (Fig. 5), which suggested that BosR was binding as an oligomeric complex. The region of DNA protected by BosR was the same in the presence or absence of Zn2+, but 10 μM Zn2+ decreased the amount of protein necessary for binding. The size of this footprint is unusually large compared with the Bacillus subtilis PerR, which protects ≈25 bp (5), but is consistent with the 50-nt E. coli OxyR footprint (16). Also, BosR binds quite a distance from the transcriptional start site (unpublished data), ≈140 bp upstream, whereas both the Bacillus subtilis PerR and E. coli OxyR bind around the –35 element, with OxyR binding extending from the –35 to the –80 region of the promoter (16). Attempts to identify other genes in the BosR regulon by probing the entire genome with the napA target sequence did not identify any additional putative BosR-binding sites.
Fig. 5.
DNase I footprint analysis of BosR. The autoradiograph of a denaturing 6% polyacrylamide gel shows the sequence of the napA P/O protected by BosR from DNase I digestion. Increasing concentrations of BosR (0, 50, 100, 500, and 1,000 nM; lanes 1–5, respectively) were incubated with labeled probe and subjected to DNase I digestion. The sequence protected by BosR is shown on the right. The numbers indicate the position from the transcriptional start site of napA. The arrow denotes an enhanced fragment.
Discussion
The cellular responses of B. burgdorferi to the reactive oxygen and nitrogen species encountered during the initial stages of mammalian infection are poorly understood. Little is known about the enzymes responsible for protecting proteins, DNA, and lipids from damage, or how the genes encoding them are regulated. To date, only two potential oxidative stress enzymes, a superoxide dismutase encoded by sodA and a Dps/Dpr homolog encoded by napA, and no putative oxidative stress regulatory proteins have been identified (www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gbb). Here we report the following: (i) the characterization of a putative oxidative stress regulatory protein, BosR; (ii) conditions that promote its interaction with the napA P/O region; (iii) the identification of its target sequence; (iv) the regulatory effects of the protein on transcriptional fusions in E. coli; and (v) the effects of ROS on in vitro binding and transcriptional regulation.
Mobility-shift DNA-binding and footprint analyses of the interaction of BosR with a 351-bp DNA fragment upstream of napA indicated that BosR was functionally distinct from PerR of Bacillus subtilis. First, mobility-shift assays demonstrated that only Zn2+ promoted efficient binding of BosR to its target sequence. Moreover, other divalent metals ions or combinations of these ions with Zn2+ had no effect on binding, whereas Mn2+ plus Zn2+ completely inhibited protein/DNA interactions. In contrast, PerR from Bacillus subtilis requires a structural Zn2+ in combination with Fe2+, Mn2+, Ni2+, or Co2+ for binding to P/Os of genes in the PerR regulon (5). Second, BosR protected a 50-nt region on the coding strand of the napA P/O that included the regions from –137 to –187 and did not include the putative PerR binding site 15 bp upstream of the actual translation start site. This is a much larger footprint than that observed for the Bacillus subtilis PerR, which protects a high-affinity region of about 25 bp very near the –10/–35 elements (5) of genes in the PerR regulon. Within this region is a 14-bp inverted repeat known as the “per” box, and all genes regulated by PerR are associated with one or more copies of this conserved sequence. To date, no distinct “BosR” box has been identified, but as additional BosR-regulated genes are identified, a distinct target sequence may become more evident. Finally, the interaction of BosR with the napA P/O sequence was slightly enhanced by the addition of t-butyl peroxide to mobility-shift assays, whereas ROS inhibits PerR/DNA binding, suggesting a different interaction between the oxidized form of each protein with its target sequence. Taken together, these data suggest that despite their sequence homology (50.7% similarity), BosR and PerR are functionally distinct.
Transcriptional fusion analysis provided a much clearer picture of the regulatory role of BosR. In E. coli cells harboring pnapAP/O-lacZ without pJAB3 or with pJAB3 without IPTG, β-galactosidase activity was very low under all conditions tested, indicating that the napAP/O-lacZ fusion alone was not efficiently transcribed in E. coli. When IPTG was added to cultures harboring pnapAP/O-lacZ and pJAB3, induction of BosR resulted in a significant increase in β-galactosidase activity, indicating that BosR exerted a positive regulatory effect on the transcriptional fusion. More importantly, the addition of t-butyl peroxide to cultures further increased β-galactosidase activity, suggesting that maximum expression was dependent on levels of ROS. Because, BosR bound DNA in either a reduced or oxidized state in mobility-shift assays and exerted a positive regulator effect on napAP/O-lacZ fusions, we propose that BosR functions as a transcriptional activator of napA.
Although there was no apparent amino acid homology (>15% similarity), BosR appeared to be functionally similar to OxyR, a transcriptional activator of H2O2-inducible genes in E. coli. For example: (i) OxyR is capable of binding DNA in either an oxidized or a reduced state, but only the oxidized form is capable of activating transcription of target genes. E. coli reporter assays indicated that BosR activates transcription under oxidizing conditions (the presence of t-butyl peroxide), but it is not known whether reduced BosR is able to activate transcription (2). Both proteins have unusually large (45–50 bp) target sequences based on DNase I footprint analysis, although OxyR target sequences are much closer to the promoters of the genes it regulates (from –35 to –80) (16) than that of BosR (3). Although BosR required Zn2+ for efficient binding, we do not believe that it is involved in sensing ROS and activating transcription. It seems more likely that oxidation/reduction of key cysteine residues (positions 153 and 156) within BosR is responsible for oxidative sensing by a mechanism similar to that described for OxyR (2). Despite the functional similarities to OxyR, it is clear that BosR represents a class of oxidative stress response regulator that has not been recognized previously.
We propose the following model for BosR. Under normal growth conditions, transcriptionally inactive, reduced BosR [BosR(red)] remains bound to its target sequence(s). As ROS levels increase within the cell, the protein becomes oxidized to a transcriptionally active form [BosR(ox)]. Because the BosR binding sequence is >100 bp upstream of the napA P/O, and it remains bound after oxidation, it seems likely that DNA bending is required to bring BosR(ox) in contact with bound RNA polymerase, as described for NIFA (the transcriptional activator for nitrogen fixation operons) in Klebsiella (17). Productive interactions of BosR(ox) with RNA polymerase promotes open complex formation and activates transcription of key oxidative stress enzymes. Once ROS are effectively eliminated from the cell, BosR(ox) is converted to BosR(red) by the thioredoxin/thioredoxin reductase complex or some unidentified reductant to down-regulate expression of BosR-dependent genes. Further characterization of BosR, its interactions with target sequences, and the identification of additional BosR-dependent genes will help delineate the regulatory mechanism(s) and more clearly define its role in the pathogenesis of B. burgdorferi.
Acknowledgments
We thank Patricia Rosa for providing B. burgdorferi strain B31 A3, James Musser and Patricia Rosa for the critical reading of this manuscript, and Anita Mora and Gary Hettrick for graphic support.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ROS, reactive oxygen species; IPTG, isopropyl β-d-thiogalactopyranoside; P/O, promoter/operator.
References
- 1.Storz, G. & Imlay, J. A. (1999) Curr. Opin. Microbiol. 2, 188–194. [DOI] [PubMed] [Google Scholar]
- 2.Zheng, M., Aslund, F. & Storz, G. (1998) Science 279, 1718–1721. [DOI] [PubMed] [Google Scholar]
- 3.Aslund, F., Zheng, M., Beckwith, J. & Storz, G. (1999) Proc. Natl. Acad. Sci. USA 96, 6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hidalgo, E., Ding, H. & Demple, B. (1997) Cell 88, 121–129. [DOI] [PubMed] [Google Scholar]
- 5.Herbig, A. F. & Helmann, J. D. (2001) Mol. Microbiol. 41, 849–859. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., Keramati, L. & Helmann, J. D. (1995) Proc. Natl. Acad. Sci. USA 92, 8190–8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowds, B. C. (1994) FEMS Microbiol. Lett. 124, 255–263. [DOI] [PubMed] [Google Scholar]
- 8.Elias, A. F., Stewart, P. E., Grimm, D., Caimano, M. J., Eggers, C. H., Tilly, K., Bono, J. L., Akins, D. R., Radolf, J. D., Schwan, T. G. & Rosa, P. (2002) Infect. Immun. 70, 2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbour, A. G. (1984) Yale J. Biol. Med. 57, 521–525. [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 11.Brenowitz, M., Senear, D. F. & Kingston, R. E. (1989) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), pp. 12.14.11–12.14.16.
- 12.Miller, J. H. (1972) in Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 13.Carroll, J. A. & Gherardini, F. C. (1996) Infect. Immun. 64, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mongkolsuk, S. & Helmann, J. D. (2002) Mol. Microbiol. 45, 9–15. [DOI] [PubMed] [Google Scholar]
- 15.Seaver, L. C. & Imlay, J. A. (2001) J. Bacteriol. 183, 7182–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tartaglia, L. A., Gimeno, C. J., Storz, G. & Ames, B. N. (1992) J. Biol. Chem. 267, 2038–2045. [PubMed] [Google Scholar]
- 17.Hoover, T. R., Santero, E., Porter, S. & Kustu, S. (1990) Cell 63, 11–22. [DOI] [PubMed] [Google Scholar]