Abstract
β-Secretase (BACE, Asp-2) is a transmembrane aspartic proteinase responsible for cleaving the amyloid precursor protein (APP) to generate the soluble ectodomain sAPPβ and its C-terminal fragment CTFβ. CTFβ is subsequently cleaved by γ-secretase to produce the neurotoxic/synaptotoxic amyloid-β peptide (Aβ) that accumulates in Alzheimer's disease. Indirect evidence has suggested that amyloidogenic APP processing may preferentially occur in lipid rafts. Here, we show that relatively little wild-type BACE is found in rafts prepared from a human neuroblastoma cell line (SH-SY5Y) by using Triton X-100 as detergent. To investigate further the significance of lipid rafts in APP processing, a glycosylphosphatidylinositol (GPI) anchor has been added to BACE, replacing the transmembrane and C-terminal domains. The GPI anchor targets the enzyme exclusively to lipid raft domains. Expression of GPIBACE substantially up-regulates the secretion of both sAPPβ and amyloid-β peptide over levels observed from cells overexpressing wild-type BACE. This effect was reversed when the lipid rafts were disrupted by depleting cellular cholesterol levels. These results suggest that processing of APP to the amyloid-β peptide occurs predominantly in lipid rafts and that BACE is the rate-limiting enzyme in this process. The processing of the APP695 isoform by GPI-BACE was up-regulated 20-fold compared with wild-type BACE, whereas only a 2-fold increase in the processing of APP751/770 was seen, implying a differential compartmentation of the APP isoforms. Changes in the local membrane environment during aging may facilitate the cosegregation of APP and BACE leading to increased β-amyloid production.
Alzheimer's disease (AD) is characterized pathologically by the formation of extracellular amyloid plaques in the brains of patients. These plaques are predominantly composed of the 4-kDa amyloid-β (Aβ) peptide, which is derived from the amyloid precursor protein (APP), a type I transmembrane protein. APP is cleaved first by β-secretase, generating the soluble ectodomain sAPPβ, and a membrane-bound C-terminal fragment (CTFβ). CTFβ is subsequently cleaved by γ-secretase to release Aβ. β-Secretase has been identified as a membrane-bound aspartic proteinase termed BACE (β-site APP-cleaving enzyme), or Asp-2 (1–3). γ-Secretase is known to be a multiprotein complex requiring presenilins for its aspartic proteinase activity (reviewed in ref. 4). A third APP-cleaving enzyme, α-secretase, cleaves within the Aβ domain releasing the soluble sAPPα domain into the extracellular space and producing CTFα, which is also a γ-secretase substrate (for review, see ref. 5).
There is increasing evidence from biochemical, epidemiological and genetic findings that cholesterol levels are linked to Aβ production and AD. The ε4 allele of apolipoprotein E, which is associated with higher plasma cholesterol levels, has been shown to be a risk factor for the disease (6), and levels of serum low-density lipoprotein and total cholesterol reportedly correlate with Aβ levels in the AD brain (7). The prevalence of AD has been shown by two independent studies to be reduced among people taking 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase inhibitors such as lovastatin (8, 9), and this finding is supported by work on APP-overexpressing mice demonstrating that increased dietary cholesterol results in higher levels of Aβ (10). Studies examining the effects of cholesterol loading and depletion on cells in culture also show a strong link between cholesterol levels and Aβ production (11, 12); however, the mechanisms connecting the two are as yet unclear.
A recent hypothesis suggests that lipid rafts could be involved in APP processing (13). Rafts are areas of the membrane rich in sphingolipids and acylated proteins, as well as cholesterol. Certain types of lipid-anchored proteins such as glycosylphosphatidylinositol (GPI)-anchored proteins (14), are associated with these rafts, and rafts have been suggested to be important in some membrane signaling and trafficking events (15). Biochemically, rafts are characterized by their insolubility in non-ionic detergents such as Triton X-100 at 4°C (16), and by this method several proteins relevant to Aβ production have been shown to be present in raft domains. These proteins include a small proportion of APP (17, 18), BACE (19), and the presenilins (20), prompting the hypothesis that amyloidogenic processing of APP may take place in lipid rafts. The putative α-secretase ADAM10, however, is predominantly soluble after detergent extraction (21), leading to a model being proposed in which amyloidogenic and non-amyloidogenic processing of APP occur in separate membrane compartments (13). According to this model, a high concentration of membrane cholesterol therefore favors Aβ production whereas a reduced cholesterol level favors the non-amyloidogenic α-secretase pathway.
To investigate the significance of lipid rafts in APP processing, we have used the approach of targeting BACE to lipid raft domains by replacing its transmembrane and cytosolic domains with a GPI anchor attachment signal. Here we show that expression of the resulting GPI-anchored form of BACE (GPIBACE) in a human neuroblastoma cell line (SH-SY5Y) substantially up-regulates both the secretion of sAPPβ into the medium and the production of Aβ, when compared with the same cell line expressing wild-type BACE. Disruption of the rafts by depletion of cholesterol reduced the formation of sAPPβ in both GPI-BACE- and wild-type BACE-expressing cells. Together, these data provide direct evidence for rafts being the cellular site of Aβ production, and that BACE is the rate-limiting step in this process.
Experimental Procedures
Generation of GPI-BACE Construct. MycHis-tagged BACE in the expression vector pcDNA3.1 (1) was stably expressed in SH-SY5Y cells. A GPI-anchored form of angiotensin converting enzyme (22) was provided by R. Skidgel (University of Illinois, Chicago) and supplied in the expression vector pIRESneo by E. Parkin (University of Leeds). The angiotensin-converting enzyme insert was cleaved from this vector by using EcoRV and NotI, leaving the pIRESneo vector containing the GPI anchor signal sequence from human carboxypeptidase M. The pro- and catalytic domains of BACE were cleaved from pcDNA3.1 by using EcoRV and BspHI, and then blunt-end ligated into pIRESneo to produce the GPI-BACE vector.
Cell Culture. SH-SY5Y cells were grown in Dulbecco's modified Eagle's medium/Ham's F-12 (1:1 mix) supplemented with 10% (vol/vol) FCS, nonessential amino acids, penicillin (50 units/ml), streptomycin (50 μg/ml), and glutamate (2 mM). Flasks were incubated at 37°C in 5% CO2. Stable transfections were carried out by using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions, and transfectants were selected by inclusion of G418 (600 μg/ml) in the medium. For analysis of secreted proteins, cells were grown to confluency, and the medium was changed to OptiMEM (Gibco/BRL). After 7–24 h of incubation, the medium was harvested and either assayed for Aβ as described below, or concentrated by centrifugation in a Vivaspin concentrator (molecular weight cutoff 10,000) and analyzed by SDS/PAGE and Western blotting. To analyze cellular proteins, cells were lysed in 50 mM Tris·HCl, pH 7.4, containing 1% Triton X-100 and a protease inhibitor mixture (Roche Diagnostics).
For the cholesterol depletion experiment, cells were treated with 4 μM lovastatin and 200 μM mevalonate in OptiMEM for 72 h. Methyl-β-cyclodextrin (MβCD, 5 mM) was then added for 10 min. Fresh OptiMEM containing 4 μM lovastatin and 200 μM mevalonate was then added to the cells for 3 h before the media were collected and concentrated as before, and lipid raft fractions were prepared from the cells.
SDS/PAGE and Western Blot Analysis. Proteins were resolved by SDS/PAGE by using 10% polyacrylamide gels, and transferred onto Immobilon P poly(vinylidene difluoride) membranes. BACE was detected by using the monoclonal 9B21 antibody (GlaxoSmithKline, Harlow, U.K.) at 1:1,000 dilution. APP and fragments were detected by using monoclonal 6E10 antibody (Signet Laboratories, Dedham, MA) at 1:1,000, polyclonal Ab54 antibody at 1:4,000, monoclonal G26 antibody at 1:1,000 (both GlaxoSmithKline) or polyclonal anti-Kunitz protease inhibitor (KPI) antibody (D. Parkinson, Sheffield Hallam University, Sheffield, U.K.) at 1:500. These four antibodies recognize residues 1–17 of Aβ, the APP C terminus, sAPPβ, and the KPI domain of APP, respectively. The antibodies against flotillin (BD Biosciences) and β-actin (Sigma) were used at 1:250 and 1:5,000, respectively. Horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch) were used at 1:1,000, and bound antibodies were detected by using the enhanced chemiluminescence system (Amersham Pharmacia).
Peptide and Protein Assays. The Aβ ELISA was carried out as described (23). Protein was quantified by using bicinchoninic acid in a microtiter plate with BSA as standard.
Phospholipase-Mediated Release of GPI-BACE. Phosphatidylinositol-specific phospholipase C (PI-PLC) purified from Bacillus thuringiensis (24) was diluted in 10 mM Hepes/NaOH, 150 mM NaCl, pH 7.4, and filter-sterilized. Confluent cells were rinsed in OptiMEM and then incubated in OptiMEM containing PI-PLC at a concentration of 0.17 mg/ml for 7 h. Media were harvested, centrifuged at 150,000 × g for 30 min, then concentrated and analyzed by SDS/PAGE and immunoblotting.
Neuraminidase and O-Glycosidase Treatment. Confluent cells were incubated in OptiMEM for 16 h, after which time the medium was collected and concentrated, and the protein concentration was equalized. Samples of media were boiled for 5 min in 250 mM sodium acetate, pH 5.5, containing 3% (wt/vol) octylglucopyranoside, 0.5% (wt/vol) SDS, and 5% (vol/vol) 2-mercaptoethanol. Neuraminidase (5 milliunits; Sigma) and 1% (wt/vol) Triton X-100 were added, and samples were incubated for 16 h at 37°C. Following incubation, 1.1 milliunits of O-glycosidase (Sigma) in 500 mM Tris maleate, pH7.5, containing 0.6% (wt/vol) octylglucopyranoside was added, and samples were incubated for a further 16 h at 37°C. Equal volumes of treated and untreated samples were resolved by SDS/PAGE.
Lipid Raft Isolation. All steps were carried out at 4°C. Confluent cells were scraped into 2 ml Mes-buffered saline (MBS, 25 mM Mes, 0.15M NaCl, pH 6.5) containing 1% (vol/vol) Triton X-100 and resuspended by passing 5 times through a 25-gauge needle. An equal volume of 90% (wt/vol) sucrose in MBS was then added. Aliquots (1 ml) were placed in 5-ml ultracentrifuge tubes, and 4-ml discontinuous sucrose gradients consisting of 35% (wt/vol) sucrose in MBS (2 ml) and 5% (wt/vol) sucrose in MBS (2 ml) were layered on top. The sucrose gradients were centrifuged at 100,000 × g for 18 h at 4°C in a Beckman SW55 rotor, and fractions (0.5 ml) were subsequently harvested from the top to the bottom of the tube.
Results
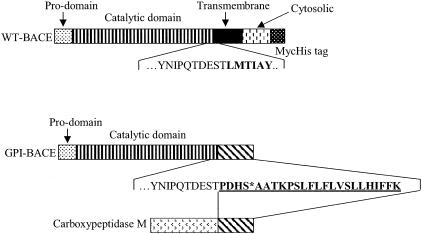
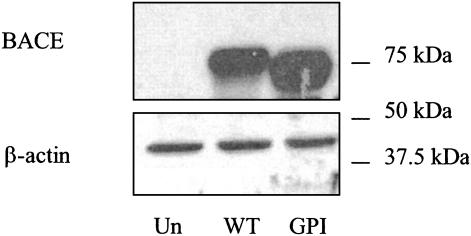
Expression of WT-BACE and GPI-BACE. Human neuroblastoma SHSY5Y cells were stably transfected with cDNA encoding either WT-BACE or GPI-BACE in which the sequence encoding the transmembrane and cytosolic domains of BACE had been replaced with the sequence encoding the GPI anchor attachment signal of carboxypeptidase M (Fig. 1). Lysates from untransfected SH-SY5Y cells and those transfected with WT-BACE or GPI-BACE were immunoblotted with a BACE monoclonal antibody and a polyclonal β-actin antibody (Fig. 2). WT-BACE migrated with a molecular mass of ≈70 kDa whereas GPI-BACE migrated at a slightly lower molecular weight due to the lack of the transmembrane and cytosolic domains. The level of expression of stably transfected WT-BACE was ≈75% that of GPIBACE.
Fig. 1.
Schematic diagram of MycHis-tagged human BACE (WT-BACE) and the GPI-anchored construct (GPI-BACE). The amino acid sequence of WT-BACE at the start of the transmembrane domain (bold) is shown. In GPI-BACE, the transmembrane, cytosolic, and MycHis domains were replaced with the 24-aa GPI anchor signal sequence (bold, underlined) of human carboxypeptidase M. The GPI attachment site is marked (*).
Fig. 2.
Expression of WT-BACE and GPI-BACE in SH-SY5Y cells. SH-SY5Y cells were stably transfected with either WT-BACE or GPI-BACE as described in Experimental Procedures. Cell lysates were prepared from transfected (WT or GPI) and untransfected (Un) cells, and immunoblotted by using the monoclonal 9B21 antibody to BACE or a polyclonal β-actin antibody.
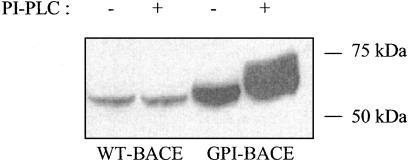
GPI-BACE Can Be Released from Cells by Treatment with PI-PLC. To confirm that GPI-BACE is attached to the cell membrane via a GPI anchor, cells stably expressing WT-BACE or GPI-BACE were treated with exogenous bacterial PI-PLC. Media from these and untreated cells were then analyzed by SDS/PAGE and immunoblotting with an antibody to the catalytic domain of BACE. In medium from WT-BACE-expressing cells, a low, but detectable, level of BACE is found both with and without PI-PLC treatment (Fig. 3). This result agrees with a previous observation that the ectodomain of the membrane-bound BACE is shed into the medium to a small extent (25). GPI-BACE also appears to undergo this shedding event, but to a greater extent (2.9 ± 0.8-fold increase) than the WT-BACE (Fig. 3). On PI-PLC treatment of GPI-BACE-expressing cells, a second band of BACE is detected in the medium at a slightly higher molecular weight. This predominant form of secreted BACE is likely to be the product of PI-PLC-mediated cleavage of the GPI anchor whereas the smaller, less abundant form of soluble BACE results from protease cleavage in the juxtamembrane region. These results demonstrate that GPI-BACE is present at the cell surface where it can be released from the membrane by the action of PI-PLC and is therefore GPI-anchored.
Fig. 3.
Release of GPI-BACE from cells by exogenous PI-PLC treatment. Confluent SH-SY5Y cells stably expressing WT-BACE or GPI-BACE were incubated in OptiMEM ± PI-PLC for 7 h, after which the medium was collected, centrifuged at 150,000 × g for 30 min, and concentrated. Equal amounts of protein from media samples were analyzed by SDS/PAGE and immunoblotted with the monoclonal 9B21 antibody to BACE.
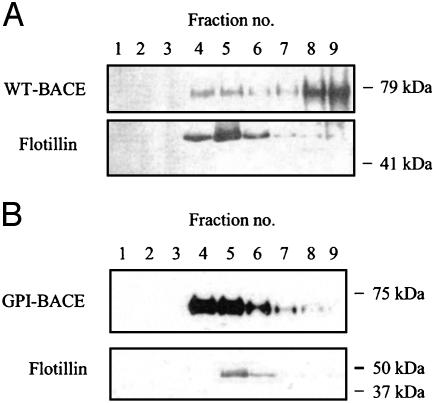
GPI-BACE Is Localized in Lipid Rafts. BACE can be detected in lipid rafts, the proportion depending on the particular detergent used for raft isolation (19). To determine whether GPI-BACE is raft-localized, cells were solubilized in Triton X-100 and separated by buoyant density gradient centrifugation. Gradient fractions were then immunoblotted with a BACE monoclonal antibody and a monoclonal antibody to flotillin, a raft-marker protein (26) (Fig. 4). All of the flotillin detected from both WT-BACE- and GPI-BACE-expressing cells was found in the raft fractions (4–6) as expected. By the Triton solubilization method used here, the majority of WT-BACE was found in the soluble, non-raft fractions (7–9), with a small proportion (≈10%) present in the raft fractions. GPI-BACE, however, was located almost exclusively in the raft fractions as expected of a GPI-anchored protein under these extraction conditions (27).
Fig. 4.
Isolation of lipid rafts from SH-SY5Y cells stably expressing WT-BACE or GPI-BACE. Lipid rafts were prepared as described in Experimental Procedures from cells stably expressing WT-BACE (A) or GPI-BACE (B). Sucrose gradients were harvested in 0.5-ml fractions (fraction 1, top of gradient; fraction 9, bottom of gradient), and each fraction was analyzed by SDS/PAGE and immunoblotted with the monoclonal 9B21 BACE antibody or a monoclonal flotillin antibody.
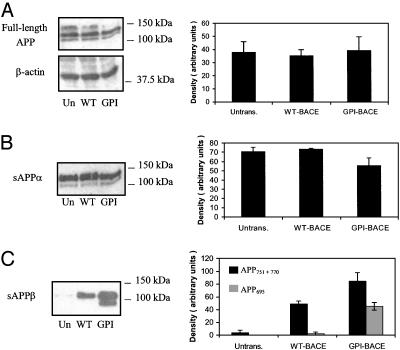
GPI-BACE Exhibits Increased APP-Cleaving Activity. It has been proposed that the amyloidogenic processing of APP by BACE and γ-secretase may take place in lipid raft domains (13). We hypothesised that, if this were the case, targeting BACE to lipid rafts by the addition of a GPI anchor should cause it to colocalize with the small fraction of APP present in rafts, substantially increasing its amyloidogenic processing. To investigate this possibility, SH-SY5Y cells stably expressing WT-BACE or GPIBACE, and untransfected cells, were grown for 16 h in serum-free medium, after which time full-length APP expression and sAPPα and sAPPβ secretion were evaluated by immunoblotting. The level of full-length APP was found to be unchanged on expression of either WT- or GPI-BACE (Fig. 5A), indicating that any changes observed in secreted APP are not due to altered APP holoprotein expression.
Fig. 5.
Comparison of APP cleavage by WT-BACE and GPI-BACE. Confluent untransfected SH-SY5Y cells or cells stably expressing WT-BACE or GPI-BACE were incubated in OptiMEM for 16 h, after which the medium was collected and concentrated and cell lysates were prepared. Equal amounts of protein from media and lysates were analyzed by SDS/PAGE and immunoblotting. (A) Detection of full-length APP and β-actin in cell lysates with the polyclonal Ab54 antibody to APP and a polyclonal β-actin antibody. (B) Detection of sAPPα in media by using the monoclonal 6E10 antibody. (C) Detection of sAPPβ in media by using antibody G26, which recognizes the epitope of APP produced by β-site cleavage. Densitometric analysis of immunoblots was carried out, and the mean results ± SEM (n = 3) are shown graphically.
The α-secretase cleavage product, sAPPα, was detected in conditioned medium by immunoblotting with the antibody 6E10, which recognizes residues 1–17 of the Aβ region. This experiment revealed a small decrease (≈10%) in secretion of sAPPα from cells overexpressing GPI-BACE when compared with levels seen from untransfected cells or cells expressing WTBACE (Fig. 5B).
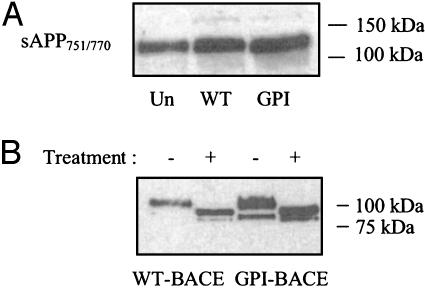
When media were analyzed by immunoblotting with an antibody recognizing the epitope produced by β-site cleavage of APP, the level of sAPPβ produced by cells expressing WTBACE was seen to be substantially increased (≈25-fold) over the low levels observed from untransfected cells (Fig. 5C). In addition, a second band of sAPPβ could be seen at a slightly lower molecular weight. On expression of GPI-BACE, a further 2-fold increase in the upper band of sAPPβ was seen over the level secreted from WT-BACE-expressing cells. The level of the lower band, however, was increased ≈20-fold (Fig. 5C).
When the same media samples were immunoblotted with an antibody recognizing the KPI domain of APP, only the upper band was detected (Fig. 6A). This finding indicates that the lower band, which was increased 20-fold on GPI-BACE expression, is derived from APP695, the shorter, non-KPI-containing isoform. Treatment of media samples with neuraminidase and O-glycosidase, to remove O-glycans, shifted both of these bands to lower molecular weights (Fig. 6B). This result supports the hypothesis that the two bands seen in Fig. 5C arise from cleavage of different isoforms of APP (APP770/751 and APP695) and not from mature and immature forms of the protein.
Fig. 6.
Analysis of sAPP isoforms secreted by WT-BACE- and GPI-BACE-expressing cells. Media were collected from untransfected SH-SY5Y cells or cells stably expressing WT-BACE or GPI-BACE as described in Fig. 5. (A) Detection of KPI-containing APP isoforms by using an anti-KPI antibody. (B) Media samples were treated with neuraminidase and O-glycosidase as described in Experimental Procedures. Treated and untreated samples were analyzed by SDS/PAGE and immunoblotting with the G26 antibody.
To determine whether the increase observed in sAPPβ production by GPI-BACE was linked to an increase in Aβ production, levels of secreted Aβ were measured by using a previously established ELISA (23). Aβ secreted from untransfected cells and those expressing WT-BACE was below the level of sensitivity of the method. However, GPI-BACE-expressing cells secreted detectable levels of both Aβ1–40 and Aβ1–42 (Table 1). Because the assay is sensitive to <0.05 ng/ml Aβ, the level of Aβ species detected from GPI-BACE-expressing cells represents an increase in Aβ1–40 of at least 11.6-fold over that seen from WT-BACE-expressing cells.
Table 1. Levels of Aβ secretion from SH-SY5Y cells stably expressing WT-BACE or GPI-BACE.
| Cell type | Aβ1-40, ng/ml | Aβ1-42, ng/ml | n |
|---|---|---|---|
| Untransfected SH-SY5Y | <0.05 | <0.05 | 3 |
| WT-BACE | <0.05 | <0.05 | 3 |
| GPI-BACE | 0.58 ± 0.03 | 0.19 ± 0.04 | 3 |
Confluent untransfected SH-SY5Y cells or cells stably expressing WT-BACE or GPI-BACE were incubated in OptiMEM for 16 h. The medium was collected and analyzed for Aβ1-40 and Aβ1-42 as described in Experimental Procedures. Results are the mean ± SEM (n = 3).
Disruption of Raft Domains by Decreasing Cellular Cholesterol Inhibits APP Cleavage by GPI-BACE. The results described above demonstrate that targeting BACE to lipid rafts increases β-cleavage of APP. To confirm that the raft-localization of GPI-BACE is essential for this up-regulation, we disrupted the raft domains by depleting cells of cholesterol and then compared levels of sAPPβ produced with those from untreated cells.
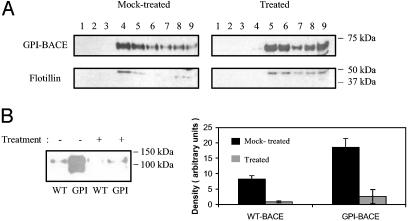
Cholesterol depletion was achieved by treatment of cells with lovastatin and MβCD. In the presence of a small amount of mevalonate, lovastatin prevents cholesterol synthesis by inhibition of 3-hydroxy-3-methylglutaryl-CoA reductase whereas MβCD extracts cholesterol from the plasma membrane. Lipid raft isolation was carried out as described in Experimental Procedures by using WT- or GPI-BACE-expressing cells incubated for equal lengths of time in the presence or absence of cholesterol-lowering agents. Fractions were analyzed by SDS/PAGE and immunoblotted for BACE and flotillin (Fig. 7A). Following treatment of the cells with lovastatin and MβCD, ≈55% of both GPI-BACE and flotillin were found in the non-raft fractions (7–9).
Fig. 7.
The effect of lovastatin and MβCD treatment on APP cleavage by GPI-BACE. Confluent SH-SY5Y cells stably expressing WT-BACE or GPI-BACE were treated with lovastatin and methyl-β-cyclodextrin as described in Experimental Procedures, or incubated in OptiMEM for equal lengths of time. The medium was collected, and lipid rafts were prepared as described. (A) Sucrose gradients were harvested in 0.5-ml fractions (fraction 1, top of gradient; fraction 9, bottom of gradient), and each fraction was analyzed for BACE and flotillin by SDS/PAGE and immunoblotting with the monoclonal 9B21 BACE antibody or a monoclonal flotillin antibody. (B) Media collected from WTBACE- or GPI-BACE-expressing cells incubated in the absence or presence of lovastatin was concentrated, and equal amounts of protein were analyzed by SDS/PAGE and immunoblotting with the G26 antibody to sAPPβ. Densitometric analysis was carried out, and the mean results ± SEM (n = 3) are shown graphically (black bars, untreated cells; gray bars, treated cells).
Media from WT-BACE- or GPI-BACE-expressing cells incubated with or without lovastatin and MβCD were collected, concentrated, and analyzed for sAPPβ (Fig. 7B). Depletion of cholesterol and the resulting disruption of lipid raft domains led to a substantial decrease in sAPPβ production from both WT-BACE- and GPI-BACE-expressing cells. Densitometric analysis showed that there was an ≈7-fold reduction in sAPPβ770/751 from both cell types (Fig. 7B) whereas sAPPβ695 production from GPI-BACE-expressing cells had been completely eliminated.
Discussion
BACE is the major, and probably only, enzyme in the brain responsible for cleavage of the amyloid precursor protein at the β-site. Knockout of BACE in mice completely abolishes production of Aβ (28, 29), and this result, together with the observation that these mice are viable, makes BACE an attractive therapeutic target. However, very little is currently known about the cellular mechanisms regulating the activity of BACE.
One major factor known to be involved in the regulation of APP processing is cholesterol. Elevated cholesterol levels have been shown to increase Aβ production via an effect on β-secretase cleavage of APP (10) whereas inhibition of cholesterol synthesis has the opposite effect (11). The exact process by which cholesterol can regulate Aβ formation is unknown, but there is increasing evidence to suggest that cholesterol-rich regions of cell membranes, known as lipid rafts, may play a part. Recent work has shown BACE to be partially raft localized when expressed in Chinese hamster ovary cells (19), and a small fraction of APP has also been shown to be present in lipid rafts in both neurons in culture (17) and mouse cerebral cortex (18).
In the present study, we provide evidence that Aβ generation in a neuronal cell line depends on lipid rafts. Riddell et al. (19) have shown that depletion of cellular cholesterol in Chinese hamster ovary cells causes BACE to dissociate from rafts, and these conditions are also known to reduce Aβ production (11). Because only a small fraction of BACE is present in rafts under normal conditions (18), we have taken a more direct approach and have targeted BACE virtually exclusively to raft domains by replacing the transmembrane and cytosolic domains of BACE with a GPI-anchor attachment sequence. The production of both Aβ and sAPPβ was dramatically increased in cells expressing GPI-BACE as compared with cells expressing WT-BACE. We have also shown that disruption of the rafts by lovastatin and MβCD treatment caused GPI-BACE to dissociate from rafts and that this treatment inhibited production of sAPPβ. This result demonstrates that expression of GPI-BACE alone is not sufficient to cause increased APP processing, and that the raft localization of the enzyme is critical for the observed increase in sAPPβ and Aβ production. GPI anchor addition to BACE could affect its intracellular distribution because the relative content of raft microdomains may vary in different subcellular compartments. However, the present results are also supported by the observation that antibody cross-linking of APP and BACE causes them to co-patch with known raft markers and also increases Aβ production (30).
As already stated, when we expressed GPI-BACE in an SHSY5Y cell line, an increase in sAPPβ production was observed with respect to the level seen in WT-BACE-expressing cells. The main band of sAPPβ visible after SDS/PAGE and immunoblotting of media was increased ≈2-fold in GPI-BACE-expressing cells. Interestingly, however, a second band at a slightly lower molecular weight was increased ≈20-fold over the low level seen from WT-BACE-expressing cells (Fig. 5C). This band was no longer visible following cholesterol depletion. Both bands could be shifted to lower molecular weights on removal of O-glycans, suggesting that they are both mature forms of APP (Fig. 6B).
APP is encoded by a single gene containing 18 exons. Alternative splicing of two of these exons results in multiple isoforms being produced, the most predominant of which are APP695, APP751, and APP770 (31–34). The two larger isoforms contain a KPI domain whereas APP770 also contains an OX.2 domain. APP695 lacks both these regions. When media from WT- or GPI-BACE-expressing cells were immunoblotted with an antibody raised against the KPI domain of APP, only the larger band of sAPPβ could be detected. It is likely, then, that the two isoforms of sAPPβ produced by GPI-BACE-expressing cells (Fig. 5C) derive from APP695 (lower band) and APP751+770 (upper band).
These results imply that, under normal circumstances, when wild-type BACE is present, most Aβ produced is derived from KPI-containing APP isoforms. This finding is consistent with a previous report showing that, when individual APP isoforms are expressed in cell lines, cells expressing the larger isoforms produce more Aβ than those expressing APP695 (35). When an increased amount of BACE is present in raft domains, however, both the KPI- and non KPI-containing isoforms are cleaved. This result could reflect a differential localization of the isoforms within the membrane, allowing APP695 and BACE to come together only when a large proportion of BACE is present in rafts.
In summary, our results show that amyloidogenic processing of APP by BACE in SHSY-5Y cells takes place far more efficiently when BACE is raft-localized, suggesting that BACE is the rate-limiting enzyme in this process, at least in this cell line. Future transgenic studies using the GPI-BACE construct would enable the pathological relevance of raft processing to be explored further.
Changes in distribution of cholesterol and lipids within cell membranes are known to occur during aging (36). It is possible, therefore, that the local membrane environment may be altered such that more BACE, and/or other enzymes or cofactors involved in amyloidogenic processing become raft-localized, and therefore aging provides a more cooperative environment for production of Aβ. Strategies such as cholesterol reduction to segregate BACE away from its substrate, APP, may therefore be therapeutically viable. GPI-BACE itself may have potential use in a screen for BACE inhibitors.
Acknowledgments
We thank Miss M. Nimick for purification of the bacterial PI-PLC. We thank the Medical Research Council of Great Britain for financial support. J.M.C. was supported by a Biotechnology and Biological Sciences Research Council Co-operative Awards in Science and Engineering Studentship.
Abbreviations: AD, Alzheimer's disease; APP, amyloid precursor protein; Aβ, amyloid β; BACE, β-site APP-cleaving enzyme; CTFα/β, C-terminal fragment of APP after α or β cleavage; GPI, glycosylphosphatidylinositol; KPI, Kunitz protease inhibitor; MβCD, methyl-β-cyclodextrin; MBS, MES-buffered saline; PI-PLC, phosphatidylinositol-specific phospholipase C; sAPPα/β, α or β cleaved ectodomain of APP.
References
- 1.Hussain, I., Powell, D. J., Howlett, D. R., Tew, D. G., Meek, T. D., Chapman, C., Gloger, I. S., Murphy, K. E., Southan, C., Ryan, D. M., et al. (1999) Mol. Cell. Neurosci. 14, 419–427. [DOI] [PubMed] [Google Scholar]
- 2.Sinha, S., Anderson, J. P., Barbour, R., Basi, G. S., Caccavello, R., Davis, D., Doan, M., Dovey, H. F., Frigon, N., Hong, J., et al. (1999) Nature 402, 537–540. [DOI] [PubMed] [Google Scholar]
- 3.Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P., Loeloff, R., et al. (1999) Science 286, 735–741. [DOI] [PubMed] [Google Scholar]
- 4.De Strooper, B. (2003) Neuron 38, 9–12. [DOI] [PubMed] [Google Scholar]
- 5.Hooper, N. M. & Turner, A. J. (2002) Curr. Med. Chem. 9, 1107–1119. [DOI] [PubMed] [Google Scholar]
- 6.Corder, E. H., Saunders, A. M., Strittmater, W. J., Schmechel, D. E., Gaskell, P. C., Small, G. W., Roses, A. D., Haines, J. L. & Pericak-Vance, M. A. (1993) Science 261, 921–923. [DOI] [PubMed] [Google Scholar]
- 7.Kuo, Y. M., Emmerling, M. R., Bisgaier, C. L., Essenburg, A. D., Lampert, H. C., Drumm, D. & Roher, A. E. (1998) Biochem. Biophys. Res. Commun. 252, 711–715. [DOI] [PubMed] [Google Scholar]
- 8.Jick, H., Zornberg, G. L., Jick, S. S., Seshadri, S. & Drachman, D. A. (2000) Lancet 356, 1627–1631. [DOI] [PubMed] [Google Scholar]
- 9.Wolozin, B., Kellman, W., Ruosseau, P., Celesia, G. G. & Siegel, G. (2000) Arch. Neurol, 57, 1439–1443. [DOI] [PubMed] [Google Scholar]
- 10.Refolo, L. M., Malester, B., LaFrancois, J., Bryant-Thomas, T., Wang, R., Tint, G. S., Sambamurti, K., Duff, K. & Pappolla, M. A. (2000) Neurobiol. Dis. 4, 321–331. [DOI] [PubMed] [Google Scholar]
- 11.Simons, M., Keller, P., De Strooper, B., Beyreuther, K., Dotti, C. G. & Simons, K. (1998) Proc. Natl. Acad. Sci. USA 95, 6460–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fassbender, K., Simons, M., Bergmann, C., Stroick, M., Lutjohann, D., Keller, P., Runz, H., Kuhl, S., Bertsch, T., von Bergmann, K., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolozin, B. (2001) Proc. Natl. Acad. Sci. USA 98, 5371–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, D. A. & Rose, J. K. (1992) Cell 68, 533–544. [DOI] [PubMed] [Google Scholar]
- 15.Simons, K. & Toomre, D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, N. M. (1999) Mol. Membr. Biol. 16, 145–156. [DOI] [PubMed] [Google Scholar]
- 17.Bouillot, C., Prochiantz, A., Rougon, G. & Allinquant, B. (1996) J. Biol. Chem. 271, 7640–7644. [DOI] [PubMed] [Google Scholar]
- 18.Parkin, E. T., Turner, A. J. & Hooper, N. M. (1999) Biochem. J. 344, 23–30. [PMC free article] [PubMed] [Google Scholar]
- 19.Riddell, D. R., Christie, G., Hussain, I. & Dingwall, C. (2001) Curr. Biol. 11, 1288–1293. [DOI] [PubMed] [Google Scholar]
- 20.Parkin, E. T., Hussain, I., Karran, E. H., Turner, A. J. & Hooper, N. M. (1999) J. Neurochem. 72, 1534–1543. [DOI] [PubMed] [Google Scholar]
- 21.Kojro, E., Gimpl, G., Lammich, S., Marz, W. & Fahrenholz, F. (2001) Proc. Natl. Acad. Sci. USA 98, 5815–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcic, B., Deddish, P. A., Skidgel, R. A., Erdos, E. G., Minshall, R. D. & Tan, F. (2000) J. Biol. Chem. 275, 16110–16118. [DOI] [PubMed] [Google Scholar]
- 23.Hussain, I., Powell, D. J., Howlett, D. R., Chapman, G. A., Gilmour, L., Murdock, P. R., Tew, D. G., Meek, T. D., Chapman, C., Schneider, K., et al. (2000) Mol. Cell. Neurosci. 16, 609–619. [DOI] [PubMed] [Google Scholar]
- 24.Low, M. G. (1992) in Lipid Modification of Proteins: A Practical Approach, eds. Hooper, N. M. & Turner, A. J. (IRL Press, Oxford), pp. 117–154.
- 25.Benjannet, S., Elagoz, A., Wickham, L., Mamarbachi, M., Munzer, J. S., Basak, A., Lazure, C., Cromlish, J. A., Sisodia, S., Checler, F., et al. (2001) J. Biol. Chem. 276, 10879–10887. [DOI] [PubMed] [Google Scholar]
- 26.Bickel, P. E., Scherer, P. E., Schnitzer, J. E., Oh, P., Lisanti, M. P. & Lodish, H. F. (1997) J. Biol. Chem. 272, 13793–13802. [DOI] [PubMed] [Google Scholar]
- 27.Parkin, E. T., Turner, A. J. & Hooper, N. M. (2001) Biochem. J. 358, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai, H., Wang, Y., McCarthy, D., Wen, H., Borchelt, D. R., Price, D. L. & Wong, P. C. (2001) Nat. Neurosci. 4, 233–234. [DOI] [PubMed] [Google Scholar]
- 29.Luo, Y., Bolon, B., Kahn, S., Bennett, B. D., Safura, B-K., Denis, P., Fan, W., Kha, H., Zhang, J., Gong, Y., et al. (2001) Nat. Neurosci. 4, 231–232. [DOI] [PubMed] [Google Scholar]
- 30.Ehehalt, R., Keller, P., Haass, C., Thiele, C. & Simons, K. (2003) J. Cell Biol. 160, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang, J., Lemaire, H. G., Unterbeck, A., Salbaum, J. M., Masters, C. L., Grzeschik, K. H., Multhaup, G., Beyreuther, K. & Muller-Hill, B. (1987) Nature 325, 733–736. [DOI] [PubMed] [Google Scholar]
- 32.Kitaguchi, N., Takahashi, Y., Tokushima, Y., Shiojiri, S. & Ito, H. (1988) Nature 331, 530–532. [DOI] [PubMed] [Google Scholar]
- 33.Ponte, P., Gonzalez-De Whitt, P., Schilling, J., Miller, J., Hsu, D., Greenberg, B., Davis, K., Wallace, W., Lieberburg, I. & Fuller, F. (1988) Nature 331, 525–527. [DOI] [PubMed] [Google Scholar]
- 34.Tanzi, R. E., McClatchey, A. L., Lamperti, E. D., Villa-Komaroff, L., Gusella, J. F. & Neve, R. L. (1988) Nature 331, 528–530. [DOI] [PubMed] [Google Scholar]
- 35.Ho, L. Fukuchi, K. & Younkin, S. G. (1996) J. Biol. Chem. 271, 30929–30934. [DOI] [PubMed] [Google Scholar]
- 36.Wood, W. G., Schroeder, F., Igbavboa, U., Avdulov, N. A. & Chochina, S. V. (2002) Neurobiol. Aging 23, 685–694. [DOI] [PubMed] [Google Scholar]