Abstract
The aerial architecture of plants is determined primarily by the pattern of shoot branching. Although shoot apical meristem initiation during embryogenesis has been extensively studied by molecular genetic approaches using Arabidopsis, little is known about the genetic mechanisms controlling axillary meristem initiation, mainly because of the insufficient number of mutants that specifically alter it. We identified the LAX PANICLE (LAX) and SMALL PANICLE (SPA) genes as the main regulators of axillary meristem formation in rice. LAX encodes a basic helix–loop–helix transcription factor and is expressed in the boundary between the shoot apical meristem and the region of new meristem formation. This pattern of LAX expression was repeatedly observed in every axillary meristem, consistent with our observation that LAX is involved in the formation of all types of axillary meristems throughout the ontogeny of a rice plant. Ectopic LAX expression in rice caused pleiotropic effects, including dwarfing, an altered pattern of stem elongation, darker color, bending of the lamina joint, absence of the midribs of leaves, and severe sterility.
Organogenesis occurs in plants throughout their lifetimes. The main axis of growth is determined by the production of two meristems: a primary shoot apical meristem (SAM) and a root meristem at embryogenesis. During postembryonic development, plants initiate a multitude of growth axes by forming new meristems called axillary meristems, which are generated in the axils of leaves and give rise to branch shoots and flowers (1). Therefore, the pattern of axillary meristem initiation and development is a key factor in determining plant architecture. Significant progress has been made on the molecular genetic analysis of SAM initiation during embryo development; however, little is known about the initiation of axillary meristems.
The development of an axillary meristem is controlled by two distinctive steps, namely, the initiation of a new meristem in the axil of a leaf and its outgrowth. Mutations that exhibit an altered pattern of axillary bud outgrowth have been described for various plant species (1), e.g., Arabidopsis auxin resistant1 (2), supershoot (3), max1, and max2 (4) and maize teosinte branched (5). On the other hand, there are only a few mutants in which the axillary meristem initiation is specifically altered. Maize barren inflorescence 2 (bif2) (6) and barren stalk1 (ba1) (7), Arabidopsis revoluta (rev) (8, 9), tomato lateral suppressor (ls) (10) and lateral suppressor of Arabidopsis (las), its cognate ortholog in Arabidopsis (11), tomato blind (bl) (12), and rice lax panicle (lax) (13) and monoculm 1(moc1) (14) are categorized in this class of mutants. The bif2 and ba1 exhibit severe suppression of all types of axillary meristems, implying that they are involved in genetic pathways controlling the general steps of axillary meristem initiation. Similarly, defects are observed in all types of lateral branches in tomato bl and Arabidopsis rev; however, the expressivity of their mutant phenotypes is relatively low, and the axillary meristems are produced in reduced numbers. In the tomato ls and rice lax, the effects are developmental stage-specific, and only a subset of the axillary meristems is affected. In ls, axillary meristem formation is completely blocked in the vegetative stage, whereas defects are subtle during reproductive development. In contrast, lax mutants represent the opposite pattern in which vegetative branching is normal but the axillary meristems are severely blocked in the mutants reproductive stage. The molecular genetic basis for this developmental stage-specific regulation of axillary meristem formation remains to be elucidated.
LS, REV, and BL have been cloned and shown to represent distinct classes of transcriptional regulators (8–10, 12, 15), whereas molecular cloning of ba1 and bif2 has yet to be reported. Recently, the rice MOC1 gene was reported to be an ortholog of the tomato LS and Arabidopsis LAS (14). Despite the progress made in the molecular cloning of genes, our knowledge of axillary meristem formation is still fragmentary. The downstream targets of these putative transcriptional regulators and their upstream regulators are not known, and the interactions between the genetic pathways are poorly understood. One obvious reason for this retardation is the insufficiency of mutants. Moreover, because the mutants are not combined in a single plant species, comprehensive genetic analysis is very difficult. Considering the number of advantages as a model species for molecular and genetic work and its importance as a major crop species, we chose rice as the model to study the molecular and genetic mechanisms controlling shoot branching.
During their development, the rice plants showed a monopodial growth pattern of shoot branching. Rice plants generate tillers as axillary meristems in the axil of leaves produced in 180° phyllotaxy during their vegetative growth phase (Fig. 1A) (16). Upon transition to the reproductive phase, the developmental program switches to the formation of a panicle, a rice inflorescence. The primary SAM produces several axillary meristems that become panicle branches. The apical meristem of the panicle branches behave as the primary SAM and sequentially produce higher-order axillary meristems. The first few meristems produced grow as secondary panicle branches, and the later meristems develop into lateral spikelets. Finally, the apical meristems of the primary and secondary panicle branches are transformed into terminal spikelets. Therefore, in the rice panicle, panicle branches and lateral spikelets are produced as axillary meristems (ref. 16 and Fig. 1 A).
Fig. 1.
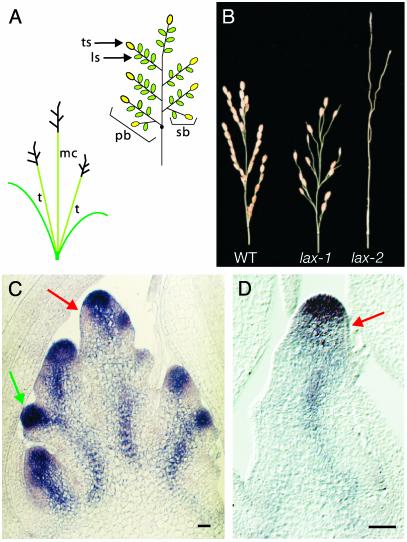
Phenotypic analysis of lax mutant. (A) A schematic of a rice plant (Left) and a panicle (Right). A rice plant is composed of a main stem called a main culm (mc) and a multiple tillers (t) produced as axillary buds. A panicle has several primary panicle branches (pb). A few secondary panicle branches (sb) and several lateral spikelets (ls) are produced on each primary panicle branch. Finally, the SAM of each panicle branch is transformed into a terminal spikelet (ts). (B) Panicle morphology of the lax mutant. WT (Left), lax-1 (Center), and lax-2 (Right) panicles. All lateral spikelets are absent, but fertile terminal spikelets are produced in lax-1, whereas formation of panicle branches and spikelets is severely reduced in lax-2.(C and D) OSH1 expression in an immature panicle at the secondary panicle branch initiation stage of WT (C) and lax-2 (D). Note: In a rice panicle, the bract leaves subtending branch shoots including panicle branches and lateral spikelets, are rudimentary and do not grow big. Red arrow shows the down-regulation of OSH1 in the site where a new meristem initiation. Green arrow shows a new axillary meristem. t, tiller; mc, main culm. (Bars = 50 μm.)
In this article, we report that the LAX and SMALL PANICLE (SPA) genes function in an overlapped genetic pathway that controls the axillary meristem initiation in rice and that the developmental stage-specific defects in lax and spa mutations are conferred by a redundancy of the two genes. We isolated the LAX gene and showed that it encodes a putative transcriptional regulator containing a basic helix–loop–helix (bHLH) domain. LAX expression was observed in the region where new axillary meristems initiate, consistent with our proposal that LAX is involved in axillary meristem initiation throughout the ontogeny of a rice plant.
Materials and Methods
Plant Materials. The lax mutant alleles analyzed in this study are shown in Table 1. The lax-1 and lax-2 alleles in Shiokari background were as described (13). lax-3 was found among a population of tissue culture-regenerated plants, and lax-4 and lax-5 were generated by 1-methyl-1-nitrosourea treatment. The spa mutant (cv. Taichung 65) was generated by ethyl methanesulfonate treatment. A lax-1 mutant with cv. Murasaki-higeyori was used for the production of lax spa double mutant.
Table 1. lax alleles analyzed in this study.
| Phenotype
|
|||||
|---|---|---|---|---|---|
| Allele | Background | Mutagen | PB | SB | LS |
| lax-1 | Shiokari* | Spontaneous | + | + | - |
| lax-2 | Shiokari | Gamma ray | ± | - | - |
| lax-3 | Nipponbare | Tissue culture | ± | - | - |
| lax-4 | Kinmaze | MNU | + | + | -† |
| lax-5 | Taichun65 | MNU | + | + | -† |
MNU, 1-methyl-1-nitorosourea; PB, primary panicle branch; SB, secondary panicle branch; LS, lateral spikelet; +, normal; ±, reduced; -, absent.
Introduced from the original lax-1 allele in Shinriki cultivars by eight crossings.
Lateral spikelets are occasionally produced in lax-4 and lax-5.
Cloning the LAX Gene. Rough mapping of the LAX locus was obtained by using cleaved amplified polymorphic sequence (CAPS) markers that were produced by using sequence information provided by the National Institute of Agrobiological Sciences. Fine mapping of the LAX locus was performed by using dCAPS markers generated according to Michaels and Amasino (17). To isolate LAX cDNA, a cDNA library made from rice inflorescence mRNA (18) was screened by using a probe amplified by PCR with primers PG1–427 (5′-CTCTCAAGCCGCCACGTGTA-3′) and PG1–1447 (5′-TGGACGAAGACACAGCAAGG-3′). The probe covered the predicted gene, PG1 (see Fig. 3A). The resultant LAX cDNA was rescued as a phagemid (pBSLAX1) as described (18).
Fig. 3.
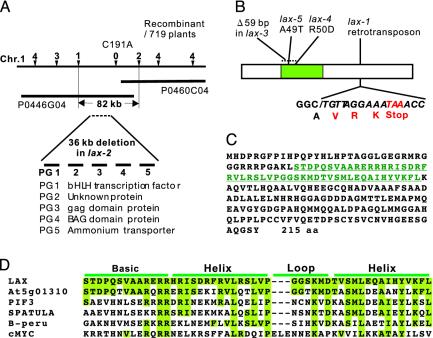
Positional cloning of the LAX gene. (A) Schematics of positional cloning. Position of the LAX locus was delimited within the 82-kb region straddling two P1 artificial chromosome clones, P0446G04 and P0460C04. Subsequently, the existence of a long deletion, which contains five predicted genes, PG1 to PG5, was identified in the lax-2 allele. (B) Lesions in lax alleles. An insertion of a retrotransposon was detected in the lax-1 allele. The sequence derived from the retrotransposon is shown in italics. A 59-bp region was deleted in lax-3. Amino acid substitutions were found in lax-4 and lax-5.(C) Amino acid sequence of LAX deduced from the cDNA sequence. The bHLH domain is shown in green. (D) Comparison of the bHLH domain of LAX and the most closely related gene of Arabidopsis (acc.At5g01310), PIF3 (AF100166), SPATULA (AF319540), maize B-Peru (S16594), and human cMYC (P23583).
Genomic DNA was amplified from lax-3, lax-4, and lax-5 mutants by using primers PG1–427 (5′-AGATTGGCCATGACGATGG-3′) and PG1–2480 (5′-AAGGAGGGCTAGCTTGATGG-3′), and the amplified fragments were purified with the QIA quick PCR Purification Kit (Qiagen Sciences, German-town, MD) and sequenced with an ABI DNA Sequencing Kit (Applied Biosystems).
In Situ Hybridization and Histological Analysis. In situ hybridization was carried out as described (18). Antisense and sense probes of LAX containing a 458–1,080 region of the LAX cDNA were generated from pBSLAX1. The antisense probe for OSH1 was generated as described (13). For histological analysis, leaves were embedded in 3% agarose, cut into 25-μm-thick sections with a microslicer, and stained with toluidine blue.
RT-PCR. For RT-PCR analysis, the first-strand cDNA was generated from random 9-mer primers (Takara Biochemicals, Ohtsu, Japan). Subsequently, the first-strand cDNA was used as a template for PCR with PG1–1116 (5′-GCCATCCACTACGTCAAGTTCC-3′) and PG1–1447 (5′-TGGACGAAGACACAGCAAGG-3′) primers. A fragment of the Actin gene was amplified from the same cDNA as a standard control to normalize the cDNA amount in the RT-PCR analysis.
Rice Transformation. Transgenic plants were produced as described (19). Ectopic expression of the LAX gene was obtained by using pBSLAX1 that had been cut by XbaI and EcoRV and subsequently cloned into the XbaI and SmaI sites of the binary vector pActnos/Hmz (20). The resultant vectors were introduced to Agrobacterium tumefaciens (strain EHA101) and used for coculture with rice calli. The Act::LAX chimeric gene was introduced to cv. Nipponbare. For the complementation test, calli were initiated from seeds harvested from WT/lax-2 plants, and the genotype of each seed was determined by PCR. Only the calli carrying the lax-2/lax-2 genotype were used for the coculture. For the PCR, PG1–1116 and PG1–1447 primers were used.
Results
LAX is required for axillary meristem generation during the reproductive phase (13) (Fig. 1B). In strong mutant alleles of the LAX locus, such as lax-2 and lax-3, the initiation of lateral spikelets is completely suppressed, and panicle branches are also severely reduced (Table 1). On the other hand, the defects were observed only in the lateral spikelets in lax-1, lax-4, and lax-5, which are weak mutant alleles of LAX.
To clarify the molecular basis of the impairment in the lax mutant, we examined the expression of OSH1, a rice KNOX gene, as a meristem marker (21). In addition to being expressed in the SAM, OSH1 is expressed in every axillary meristem, as are KNOX genes in other species (22) (Fig. 1C). In the normal development of axillary meristem initiation, OSH1 expression is down-regulated in SAM-derived cells that are destined to differentiate to bracts. Then, the OSH1 mRNA reappears in cells in the axil of the bract primordium, and these cells will develop to form the axillary meristem. Whether these cell are initiated de novo from differentiated cells (de novo initiation model) or derived from a portion of the primary SAM (detached meristem model) is still a matter of discussion (1, 23, 24). Once a new meristem is initiated, OSH1 expression reappears. In lax-2, OSH1 expression in the primary SAM and its down-regulation were observed. Initiation of OSH1 expression in new meristems, however, was not detected (Fig. 1D), suggesting that LAX specifically controls new meristem initiation and/or maintenance.
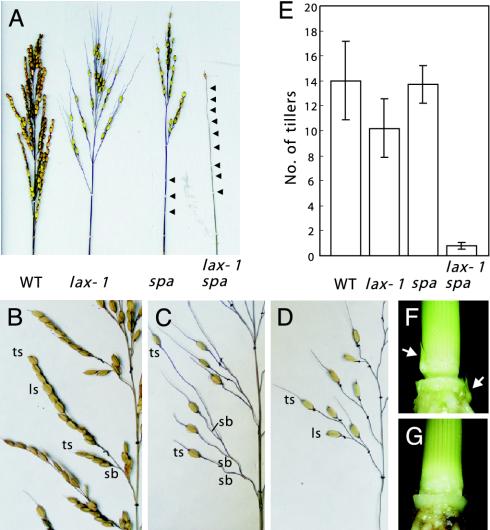
Besides lax, we isolated another panicle branching mutant, small panicle (spa). In spa, the number of panicle branches and spikelets in a panicle was reduced (Fig. 2 A–D). In particular, abolishment of primary panicle branches at lower nodes was prominent. Each primary branch was short in length, as shown in Fig. 2D, and secondary branches were rarely produced. Whereas lateral spikelets were completely abolished in the lax-1 mutant, they were generated in the spa but their number was reduced (Fig. 2 A). The lax and spa mutations exhibited dramatic synergestic effects on axillary meristem initiation. When the spa mutation was combined with even a weak allele of lax, lax-1, in which only lateral spikelets are affected, the panicle became a wire-like structure devoid of all branches (Fig. 2 A). Furthermore, although the lax or spa single mutants showed little effect on tillering, vegetative shoot branching was almost completely suppressed (Fig. 2 E–G). The phenotype of the double mutant clearly demonstrated that the formation of all axillary meristems in rice is under the control of a single genetic mechanism, in which LAX and SPA play crucial roles.
Fig. 2.
Phenotypic analysis of spa and lax spa double mutants. (A) Comparison of the panicle phenotypes of the lax, spa single mutants, and the lax spa double mutant. Arrowheads indicate panicle nodes. (B–D) Close-up views of panicles in WT (B), lax-1 (C), and spa (D) mutants. (E) Effects of lax and spa mutations on the number of tillers, vegetative stage branching. Number of panicles, except for one on a main culm, are shown as the number of tillers. Number of plants examined were 68, 42, 136, and 6 for WT, lax-1, spa, and lax-1 spa, respectively. (F and G) Tiller buds of vegetative stage plants. Arrows indicate tiller buds in the WT plant (F). Tiller buds were not observed in lax spa double mutant plants (G).
The LAX locus was previously mapped to the long arm of chromosome 1 (25). We used a population of 719 F2 plants obtained from a cross between Japonica (lax-2/lax-2) and Indica (WT/WT) and delimited the LAX region to within 82 kb (Fig. 3A). Then, we found a long deletion in the lax-2 allele spanning >36 kb, which contained five predicted genes, PG1 to PG5. Sequencing these five genes in the lax-1 allele resulted in the detection of a retrotransposon insertion in PG1, whereas no mutations were found in the other four predicted genes. Sequence alterations were also identified in PG1 in the other lax alleles, suggesting that PG1 likely represents the LAX gene (Fig. 3B). To obtain additional evidence of this, PG1 was introduced into lax-2 plants under the control of a rice Actin promoter that confers strong and constitutive expression in rice (26). Five independent transgenic lines were generated, and three plants showed a rescued panicle phenotype and additional effects caused by ectopic LAX expression (described below). See Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org, for a view of a panicle of a lax-2/lax-2;ACT::LAX/+ plant. Taking all of these findings together, we concluded that PG1 is the LAX gene.
We obtained a 1,080-bp LAX cDNA clone by screening a cDNA library prepared from very young inflorescences (18). Sequencing of the LAX cDNA revealed that the LAX gene is intronless and encodes an ORF of 215 aa (Fig. 3C). A bHLH domain was identified in its amino acid sequence, suggesting that LAX is a transcriptional regulator. The LAX bHLH domain showed high sequence similarity to that of other plant bHLH proteins and bHLH proteins predicted from the rice genome sequence (Fig. 3D and Fig. 7, which is published as supporting information on the PNAS web site). Outside of the bHLH region, however, no other conserved domains were identified. A database search failed to identify cognate homologs of LAX in the Arabidopsis genome, suggesting a possibility that LAX represents a grass-specific regulator of shoot branching.
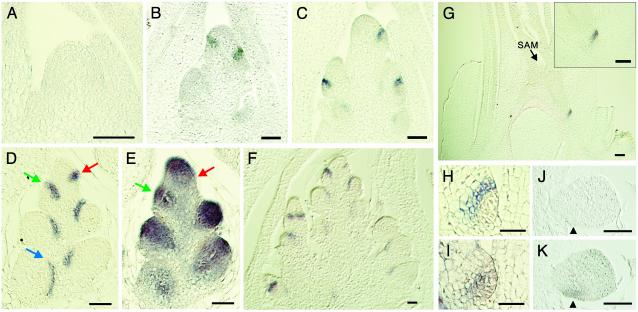
The expression pattern of LAX was examined by in situ hybridization. LAX expression was not detected in the vegetative SAM (Fig. 4A). After transition to the reproductive phase, LAX mRNA was detected in a layer of one or two cells in width (Fig. 4 B and C). To reveal the timing of LAX expression more precisely, we compared LAX and OSH1 expressions in consecutive sections. The analysis showed that the layer where LAX mRNA accumulated corresponds to the boundary between the new meristem and the SAM (Fig. 4 D and E). Although this tissue layer was carefully examined, no discernable structures were apparent in the layer, nor are we aware of any published evidence for distinct structure in this position. The earliest LAX mRNA expression was observed when down-regulation of OSH1 in the incipient shoot branches occurred (Fig. 4 D and E, red arrow). The LAX expression was maintained after a new meristem started to grow (Fig. 4 D and E, green arrow), but it gradually disappeared as the new meristem elongated (Fig. 4D, blue arrow). This pattern of LAX expression was repeatedly observed in every axillary meristem, including secondary panicle branches and lateral spikelets (Fig. 4F). When a primordium of a tiller shoot initiated, LAX mRNA was detected in a few cell layers above the area surrounding the new meristem (Fig. 4G). Expression of LAX in axillary meristems, which will give rise to tiller buds, is consistent with the lax spa double-mutant phenotype. In accordance with the normal development of the primary SAM in the lax spa double mutant, LAX expression was not detected during embryogenesis (Fig. 4 J and K). No signal of LAX mRNA accumulation was observed in leaves, stems (Fig. 4 B, C, and G), or roots (Fig. 8, which is published as supporting information in the PNAS web site).
Fig. 4.
Expression pattern of the LAX gene. RNA in situ hybridization of LAX (A–D, F–H, and J) and OSH1 (E, I, and K). (A) Longitudinal section of a vegetative SAM. (B–F) Longitudinal section of an inflorescence meristem at the primary branch differentiation stage (B), the secondary branch differentiation stage (C), and the spikelet initiation stage (D–F). LAX mRNA was specifically expressed in the upper-boundary layer between the meristem and the region in which the axillary meristem will develop. Comparison of the expression pattern of LAX (D) and OSH1 (E) in consecutive sections shows that LAX expression has been initiated where OSH1 down-regulation took place (red arrow). When the new axillary meristem started to grow, both LAX and OSH1 were expressed (green arrows). (F) The same expression pattern of LAX was repeatedly observed in the places where new axillary meristems initiate. (G) LAX mRNA was expressed in the tiller primordium. (H and I) LAX and OSH1 mRNA accumulation during tiller bud initiation. Analysis of consecutive sections indicated that LAX mRNA localization (H) is in two cell layers in the upper region of OSH1 expression (I). (J and K) LAX and OSH1 expressions in the late globular-stage embryo. OSH1 expression was observed in the region where a primary SAM initiates (arrowhead), whereas the signal of LAX expression was not observed. (Bars = 50 μm.)
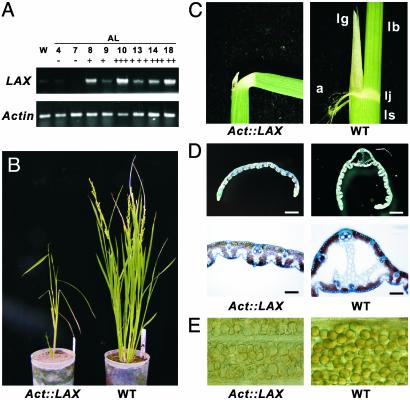
To obtain further insights into the LAX function, we produced transgenic rice plants expressing LAX under the control of a strong and constitutive Actin promoter (26). Abnormal development was observed in 13 independent transgenic plants among 19 plants generated. Although the severity varied among the 13 plants, all showed the same phenotypic spectrum. Overexpression of LAX was confirmed by RT-PCR analysis (Fig. 5A). A clear correlation between the severity of phenotypes and the level of LAX expression strongly suggested that the observed phenotypes were indeed caused by the ectopic expression of LAX (Table 2, which is published as supporting information on the PNAS web site). A representative plant is shown in Fig. 5B. Act::LAX plants showed pleiotropic effects, including dwarfing, an altered pattern of stem elongation, darker color, bending of the lamina joint, absence of the midribs of leaves, and severe sterility (Fig. 5 C–E). To our surprise, the pattern of axillary meristem initiation and morphology of the panicle and spikelet were not significantly altered. Among the abnormalities found in the Act::LAX plants, lamina-joint bending was of particular interest because it mimicked the response of rice leaves to exogenously applied auxin or Brassinosteroide (27, 28). This finding raises the possibility that ectopic expression of LAX disturbed the signal transduction pathway of these hormones.
Fig. 5.
Phenotypes of Act::LAX transgenic rice plants. (A) Ectopic expression of LAX in leaves of Act::LAX transgenic plants. The + indicates severity of the phenotype. (B) A representative Act::LAX transgenic rice plant (Left) and a WT plant (Right). (C) Lamina joint in an Act::LAX plant (Left) and a WT plant (Right). (D) Transverse section of leaf blades in an Act::LAX plant (Left) and WT plant (Right). The development of a midrib was observed in the WT leaf but not in the Act::LAX leaf. (E) Pollen grains in an Act::LAX transgenic plant (Left) and a WT plant (Right). Pollen in the transgenic plants look empty and shrunken. lg, ligule; lb, leaf blade; ls, leaf sheath; a, auricle; lj, lamina joint.
Discussion
LAX and SPA Play Overlapped Functions in Axillary Meristem Initiation. The molecular genetic mechanisms controlling axillary meristem initiation are relatively poorly understood as compared with that governing the primary SAM formation during embryogenesis. In many species, the pattern of axillary meristem formation depends on the developmental stage of the SAM even though the molecular basis of the developmental stage-specific regulation remains unknown. For example, in the tomato plant, axillary meristems are produced in a monopodial manner in the vegetative stage, which changes to sympodial growth after the transition to the reproductive phase (1). In the tomato ls mutant, axillary meristems are affected only in the vegetative stage, and inflorescences develop normally. This finding suggests that LS is involved in the vegetative stage-specific pathway for axillary meristem initiation. In contrast, vegetative branching is normal, but axillary meristem formation is severely suppressed in the reproductive development in the lax of rice (ref. 13 and this study). Maize lg2 mutants also show the reduction of tassel branches in a developmental stage-dependent manner (29). Here, we demonstrated that the stage-specific defects in the lax mutation are caused by redundancy with another branching gene, SPA. Interestingly, the dependence of their functions is also developmental stage-dependent. During the vegetative stage, loss of either the LAX or SPA function is fully compensated by one or the other; however, lax or spa single mutations exhibit distinctive reproductive phenotypes in the panicle.
The bHLH transcription factors, which are conserved in all eukaryotes, are important regulatory components of transcriptional networks in many developmental pathways (30. 31). Although the function of bHLH proteins has been well analyzed in animals, few plant bHLH genes have been studied in detail. Because bHLH proteins work as dimmers, a possibility that SPA is a dimerization partner of LAX and also encodes a bHLH protein is raised. However, because a large number of bHLH genes are present in plant genomes, and transcription factors of other classes, such as Myb are also possible candidates for partners of bHLH proteins (32), the anticipation of the SPA sequence from genome database seems impractical. The molecular cloning of SPA, which is currently underway, is a prerequisite to obtaining further insights into the genetic framework controlling reproductive branching in rice.
LAX and SPA Are General Regulators of Shoot Branching in Rice. Our results also indicated the existence of a general regulatory mechanism that governs the formation of all axillary meristems, irrespective of the future identity of the branch shoots. Obviously, this pathway is independent of that controlling the primary SAM formation during embryogenesis because the development of the SAM is normal even in the lax spa double mutant. This notion was well supported by the LAX expression pattern, in which LAX mRNA was detected in all axillary meristems throughout postgermination ontogeny, but not during SAM formation during embryogenesis. Recently, it was reported that rice moc1 also exhibits severe defects in all axillary meristems, indicating that MOC1 is also a general regulator of the axillary meristem initiation in rice (14). Analysis of genetic and molecular interactions among MOC1, LAX, and SPA will be important to establish the genetic framework for the axillary shoot branching of rice. Maize ba1 and bif2 and tomato bl mutants exhibit defects in all types of axillary meristems, suggesting that there are similar general pathways of shoot branching in maize and tomato. Based on the genetic analysis of ba1 and bif2 in maize, bl and ls in tomato act in independent pathways, suggesting there are multiple genetic pathways for axillary meristem formation (6, 12).
LAX Functions in a Non-Cell-Autonomous Manner. The mutant phenotype indicates that the site of LAX function is the region where new meristems initiate; however, LAX mRNA accumulates in a boundary between the SAM and the place where a new meristem initiates. How LAX accomplishes its noncell-autonomous function is currently unknown. One scenario is that LAX enhances or amplifies a meristem-producing signal that is generated in the SAM and transmitted to cells destined to form a new lateral meristem. It is also possible, however, that the boundary-like expression of LAX excludes an inhibitory signal to ensure the initiation or maintenance of a new meristem. However, although the expression of LAX mRNA is indicative, the possibility that the LAX protein moves to the site for new meristem formation, as demonstrated for a number of genes involved in plant development, is also worthy of consideration (33, 34).
Similar patterns of the boundary-like mRNA accumulation were reported for several genes, such as LATERAL ORGAN BOUNDARIES (LOB) (35), LAS of Arabidopsis (11), CUPSHAPED COTYLEDON (CUC)/NO APICAL MERISTEM (NAM) (36, 37), and BRANCHED SILKLESS1 (BD1)/FRIZZY PANICLE (FZP) (38, 39). The function of LOB protein is unknown, whereas LAS, CUC1/CUC2/NAM, and BD1/FZP belong to different classes of transcriptional regulators involved in the developmental regulation. The fact that a number of different genes incidentally exhibit common conspicuous patterns of mRNA expression in the boundary between the SAM and a new organ primordium suggests that the cells in the boundary may have specific identity. The BD1 and FZP genes control floral meristem identity in maize and rice, respectively. Their primary function has been proposed to be the repression of the axillary meristem initiation, which ensures the transition to the floral meristem identity. The BD1 and FZP are specifically expressed in the axil of the glume from where an ectopic axillary meristem arises in the bd1 and fzp mutants, supporting an idea that the cells in the axil contain a distinctive identity and are important for proper developmental progression.
LAX Is a Grass-Specific bHLH Family Transcription Factor. Although a number of bHLH proteins showing high sequence similarity with LAX can be found in plant proteins, the sequence conservation was limited to the bHLH domain and did not extend outside the domain, even among proteins predicted from the genome sequence of Arabidopsis. Similarly, the BD1 and FZP do not have orthologs in the Arabidopsis genome; however, their orthologs can be found in a variety of grass species and exhibit high similarity throughout the amino acid sequence, implying an intriguing possibility that they evolved to establish grass-specific developmental systems. In fact, completion of the genome sequencing of Arabidopsis and rice revealed that a certain portion of rice genes does not have homologs in Arabidopsis (40, 41). Although it is likely that LAX plays a grass-specific role in the axillary meristem formation, the function of MOC1, LS, and LAS seems to be well conserved between rice, a grass species, and tomato and Arabidopsis, dicot species, irrespective of the evolutional distance between these species (10, 11, 14). In the near future, the isolation of more genes controlling the developmental pathways in grass species will provide further insight into the mechanisms whereby grasses established their unique developmental programs by using genetic mechanisms common to grasses and other plant species and evolving novel systems unique to grasses.
Supplementary Material
Acknowledgments
We thank Y. Nagato, H. Kitano, and H. Satoh for providing the plant materials, M. Matsuoka for providing the OSH1 cDNA clone, and M. Nobuhara and S. Kohashi for their excellent technical support. Information on cleaved amplified polymorphic sequence markers was kindly provided by M. Yano. We also thank R. Schmidt for his helpful suggestions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SAM, shoot apical meristem; bHLH, basic helix–loop–helix.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB115668).
References
- 1.Grbri', V. (2001) in Meristematic Tissues in Plant Growth and Development, eds. McManus, M. T. & Veit, B. E. (Sheffield Academic, Sheffield, U.K.), pp. 142–171.
- 2.Stirnberg, P., Chatfield, S. P. & Leyser, H. M. O. (1999) Plant Physiol. 121, 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tantikanjana, T., Yong, J. W. H., Letham, D. S., Griffith, M., Hussain, M., Ljung, K., Sandberg, G. & Sundaresan, V. (2001) Genes Dev. 15, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stirnberg, P., vande Sande, K. & Leyser, H. M. O. (2002) Development (Cambridge, U.K.) 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- 5.Doebley, J., Stec, A. & Hubbard, L. (1997) Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- 6.McSteen, P. & Hake, S. (2001) Development (Cambridge, U.K.) 128, 2881–2891. [DOI] [PubMed] [Google Scholar]
- 7.Ritter, M. K., Padilla, C. M. & Schmidt, R. J. (2002) Am. J. Bot. 89, 203–210. [DOI] [PubMed] [Google Scholar]
- 8.Talbert, P. B., Adler, H. T., Parks, D. W. & Comai, L. (1995) Development (Cambridge, U.K.) 121, 2723–2735. [DOI] [PubMed] [Google Scholar]
- 9.Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N. & Clark, S. E. (2001) Plant J. 25, 223–236. [DOI] [PubMed] [Google Scholar]
- 10.Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G. & Theres, K. (1999) Proc. Natl. Acad. Sci. USA 96, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greb, T., Clarenz, O., Schäfer, E., Müller, D., Herrero, R., Schmitz, G. & Theres, K. (2003) Genes Dev. 17, 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz, G., Tillmann, E., Carriero, F., Fiore, C., Cellini, F. & Theres, K. (2002) Proc. Natl. Acad. Sci. USA 99, 1964–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu, M., Maekawa, M., Shimamoto, K. & Kyozuka, J. (2001) Dev. Biol. 231, 364–373. [DOI] [PubMed] [Google Scholar]
- 14.Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G., Zeng, D., Wang, X., Liu, X., Teng, S., Hiroshi, F., et al. (2003) Nature 422, 618–621. [DOI] [PubMed] [Google Scholar]
- 15.Zhong, R. & Ye, Z.-H. (1999) Plant Cell 11, 2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshikawa, K. (1989) The Growing Rice Plant (Nosan Gyoson Bunka Kyokai, Tokyo).
- 17.Michaels, S. D. & Amasino, R. M. (1998) Plant J. 14, 381–386. [DOI] [PubMed] [Google Scholar]
- 18.Kyozuka, J., Konishi, S., Nemoto, K., Izawa, T. & Shimamoto, K. (1998) Proc. Natl. Acad. Sci. USA 95, 1979–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. (1994) Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- 20.Sentoku, N., Sato, Y. & Matsuoka, M. (2000) Dev. Biol. 220, 358–364. [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka, M., Ichikawa, H., Saito, A., Tada, Y., Fujimura, T. & KannoMurakami, Y. (1993) Plant Cell. 5, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, D., Veit, B. & Hake, S. (1994) Development (Cambridge, U.K.) 120, 405–413. [Google Scholar]
- 23.Grbi', V. & Bleecker, A. B. (2000) Plant J. 21, 215–223. [DOI] [PubMed] [Google Scholar]
- 24.Long, J. & Barton, M. K. (2000) Dev. Biol. 218, 341–353. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita, T. & Takahashi, M. (1991) J. Fac. Agr. Hokkaido Univ. 65, 1–61. [Google Scholar]
- 26.Zang, W., McElroy, D. & Wu, R. (1991) Plant Cell 3, 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda, E. (1965) Physiol. Plantarum 18, 813–827. [Google Scholar]
- 28.Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H. & Matsuoka, M. (2000) Plant Cell 12, 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh, J. & Freeling, M. (1999) Plant J. 19, 489–495. [DOI] [PubMed] [Google Scholar]
- 30.Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B. & Bailey, P. C. (2003) Mol. Biol. Evol. 20, 735–747. [DOI] [PubMed] [Google Scholar]
- 31.Murre, C. & Massari, M. E. (2000) Mol. Cell. Biol. 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. M. & Schiefelbein, J. (1999) Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- 33.Lucas, W. J., Bouche-Pillon, S., Jackson, D. P., Nguyen, L., Baker, L., Ding, B. & Hake, S. (1995) Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- 34.Sessions, A., Yanofsky, M. F. & Weigel, D. (2000) Science 289, 779–782. [DOI] [PubMed] [Google Scholar]
- 35.Shuai, B., Reynaga-Pena, C. G. & Springer, P. S. (2002) Plant Physiol. 129, 746–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aida, M., Ishida, T. & Tasaka, M. (1999) Development (Cambridge, U.K.) 126, 1563–1570. [DOI] [PubMed] [Google Scholar]
- 37.Souer, E., van Houwelingen, A., Kloos, D., Mol, J. & Koes, R. (1996) Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- 38.Chuck, G., Muszynski, M., Kellogg, E., Hake, S. & Schmidt, R. J. (2002) Science 298, 1238–1241. [DOI] [PubMed] [Google Scholar]
- 39.Komatsu, M., Chujo, A., Shimamoto, K. & Kyozuka, J. (2003) Development (Cambridge, U.K.) 130, 3841–3850. [DOI] [PubMed] [Google Scholar]
- 40.Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. (2002) Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 41.Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002) Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.