Abstract
RAR1 and its interacting partner SGT1 play a central role in plant disease resistance triggered by a number of resistance (R) proteins. We identified cytosolic heat shock protein 90 (HSP90), a molecular chaperone, as another RAR1 interacting protein by yeast two-hybrid screening. RAR1 interacts with the N-terminal half of HSP90 that contains the ATPase domain. HSP90 also specifically interacts with SGT1 that contains a tetratricopeptide repeat motif and a domain with similarity to the cochaperone p23. In Arabidopsis, the HSP90 inhibitor geldanamycin reduces the hypersensitive response and abolishes resistance triggered by the R protein RPS2 against Pseudomonas syringae pv. tomato DC3000 (avrRpt2). One of four Arabidopsis cytosolic HSP90 isoforms, AtHSP90.1 is required for full RPS2 resistance and is rapidly induced upon pathogen challenge. We propose that RAR1 and SGT1 function closely with HSP90 in chaperoning roles that are essential for disease resistance.
Eukaryotes are continually attacked by microorganisms with sophisticated strategies to colonize their hosts. In plants, pathogen infection is countered by a surveillance system consisting of resistance (R) proteins that detect corresponding pathogen determinants (1). Resistance responses activated by various R proteins often include rapid ion fluxes, generation of reactive oxygen species (ROS), and production of antimicrobial compounds. These responses are often accompanied by localized programmed cell death, the hypersensitive response (HR), at the site of pathogen invasion (2). The universal response patterns and the structural similarities shared by R proteins suggest that common signal transduction pathways may be used upon pathogen recognition. Moreover, R proteins share striking similarities with components of the animal innate immune system, suggesting that some downstream signaling components may be common to both plants and animals (3).
RAR1 (required for Mla12 resistance) is an essential component of resistance conferred by many R genes (4–6). In barley, rar1 mutants fail to accumulate ROS or mount the HR (4). Similarly, Arabidopsis rar1 mutants are defective in R-protein mediated resistance against several bacterial (Pseudomonas) and oomycete (Peronospora) pathogens, highlighting the importance of RAR1 in disease resistance (5, 6). However, the precise biochemical function of this protein remains unclear. RAR1 contains two zinc-binding modules termed cysteine- and histidine-rich domain (CHORD)-I and CHORD-II, which are characterized by six conserved cysteines and three conserved histidines (4). CHORD-containing proteins have been found in all tested eukaryotes except yeast, and the conserved primary structure of CHORD domains and their tandem organization suggests that these proteins serve important cellular functions. RNA silencing of the gene encoding the Caenorhabditis elegans CHORD protein results in embryo lethality and gonad hyperplasia, indicating an involvement of RAR1 homologs in animal development (4). An animal RAR1 homolog, Melusin, originally identified as a β-integrin interacting protein in the yeast two-hybrid system, is expressed in heart muscle, where it is required for sensing stress from pressure overload (7). The variety of biological systems in which CHORD proteins are involved suggests that RAR1 may be a component of conserved processes central to many cellular activities.
RAR1 interacts with a conserved protein, SGT1 (suppressor of the G2 allele of skp1), that was originally identified as an essential component of cell cycle control in yeast (8, 9). Mutation analysis in Arabidopsis and gene silencing experiments in barley and Nicotiana benthamiana demonstrated that SGT1 is required for disease resistance mediated by diverse R proteins (9–13). In yeast, Sgt1 physically interacts with Skp1 and activates assembly of the centromere-binding factor 3 (CBF3) kinetochore complex (8). There, Sgt1 is also required for the function of an SCF (Skp1–Cul1–F box) ubiquitin ligase complex that mediates ubiquitylation and degradation of Sic1, an inhibitor of Cdc28 kinase (8). Importantly, plant and human SGT1 genes can complement cell-cycle defects in yeast sgt1 mutants, suggesting that the biochemical activity of SGT1 is highly conserved in eukaryotes (8, 9). Consistent with this idea, Arabidopsis SGT1b is required for the auxin response mediated by an SCF complex (14). Moreover, in plants, SGT1 interacts with the SCF complex subunits SKP1 and CUL1 and associates with the COP9 signalosome that regulates the SCF complex by removing the small ubiquitin-like protein RUB1 from CUL1 (9, 12). Silencing SCF and COP9 signalosome components in plants compromises virus resistance, indicating the importance of ubiquitylation in disease resistance (12).
To further understand how RAR1 and SGT1 function in the resistance pathways, we searched for additional RAR1-interacting proteins by yeast two-hybrid screening. Here we report that cytosolic heat shock protein 90 (HSP90), a molecular chaperone, specifically interacts with both RAR1 and SGT1. We demonstrate that the specific HSP90 inhibitor geldanamycin (GDA) inhibits the HR cell death and resistance mediated by the Arabidopsis R protein RPS2. Furthermore, mutations in an Arabidopsis cytosolic HSP90 isoform attenuate RPS2-mediated resistance and HR cell death, indicating that HSP90 is an essential factor for the resistance response. Together, these data suggest that RAR1 and SGT1 may function as cochaperones of HSP90 in processes essential for plant disease resistance.
Materials and Methods
Plant Materials. The barley (Hordeum vulgare) cultivar Sultan5 containing either wild-type HvRAR1 or hvrar1–2 has been described (9). Barley plants were grown at 20°C with 16 h of light and 8 h of darkness. The Arabidopsis AtHSP90.1 T-DNA insertion lines in Col-0 ecotype (contains RPM1 and RPS2) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Insertion mutant information was obtained from The Salk Institute Genomic Analysis Laboratory web site (http://signal.salk.edu). The T-DNA insertion sites were confirmed by PCR using T-DNA left border primer 5′-GCG TGG ACC GCT TGC TGC AAC T-3′ and AtHSP90.1-specific primer 5′-CTT AGC TTG TGC TCG ATC TTC-3′. Arabidopsis growth conditions have been described (6).
Pathogen Strains and Pathology Tests. Pseudomonas syringae pv. tomato (Pst) DC3000 strains were grown overnight with kanamycin (25 μg/ml) and rifampicin (100 μg/ml) as described (15), washed once in 10 mM MgCl2, and resuspended to a density of 1 × 105 colony-forming units (cfu)/ml for in planta growth assays and 1 × 107 cfu/ml for HR tests. Bacterial suspensions were infiltrated into abaxial leaf surfaces by using a needleless syringe, and the HR was detected by trypan blue staining as described (15, 16). Bacterial growth tests were performed as described (15). For GDA (Sigma) inhibition experiments, 10–50 μM of GDA (diluted from 10 mM stock in DMSO) or an equivalent concentration of DMSO alone as control was applied into leaves by infiltrating concomitantly with Pst DC3000 strains.
Screening and Interaction Assays Using Yeast Two Hybrid. The Arabidopsis cDNA library, RAR1 and SGT1 clones, the yeast two hybrid screening methods and interaction assays were described previously (9). The barley cDNA library was created in the pB42AD vector (Clontech) by using poly(A)+ RNA isolated from barley leaf tissue (cultivar Sultan5) infected with powdery mildew (Blumeria graminis f. sp. hordei) incompatible isolate A6 (gift from P. Piffanelli and P. Schulze-Lefert, The Sainsbury Laboratory). The full-length barley HSP90 (HvHSP90) cDNA (GenBank accession no. AY325266) was isolated from the barley cDNA library by using HvHSP90 specific primer, 5′-CCA GCA GAA CAA GAT CCT CAA GG-3′ and pB42AD vector primer 5′-CTG GTT CAG AAT TGC TGC AGG TCG-3′. The deletion constructs, HvHSP90-Δhalf (amino acids 1–413) and HvSGT1-int (amino acids 115–294), were created by using an internal StuI site, and the HvSGT1 clone RNAi-SGT1 described earlier (9), respectively.
In Vitro Protein-Binding Assay. A plasmid expressing HvRAR1 as an N-terminal S-tag and C-terminal His-tag fusion protein was constructed by using unique BamHI and HindIII sites within pSTAG (4). The S-HvRAR1-His fusion protein was purified by using a His-bind resin as instructed by the supplier (Novagen). The S-HvCHORD-I and S-HvCHORD-II fusion proteins were purified as described (4). Human Hsp90 (HsHsp90) protein was purchased from StressGen Biotechnologies (Victoria, BC, Canada). Approximate molar equivalents of each of the proteins were mixed in the binding buffer (45 mM Tris·HCl, pH 7.5/50 mM NaCl/10 mM KCl/3 mM MgCl2/0.1% Nonidet P-40, modified from ref. 17) and incubated for 30 min at 25°C. S-protein agarose beads (Novagen) for precipitating S-tag fusion proteins were added to the mixture and incubated for 2 h at 4°C. Beads were washed, and proteins were eluted and analyzed by SDS/PAGE. HsHsp90 and HvRAR1 fusion derivatives were visualized by HsHsp90-specific antibody (StressGen Biotechnologies) and RAR1 antibody (9), respectively.
Antibody Production and Immunoblot Analysis. The fragment containing the N-terminal half of HvHSP90 was subcloned from the pB42AD vector into pSTAG by using EcoRI and HindIII sites. The resulting S-tag fusion protein was overexpressed in Escherichia coli BL21(DE3) (pLysS, pSBET), purified and used for raising anti-HvHSP90 antibodies in rats as described (9). Anti-RAR1 and anti-SGT1 antibodies as well as methods for coimmunoprecipitation were described (9).
AtHSP90.1 Expression Analysis. Wild-type Arabidopsis Col-0, athsp90.1-1, or athsp90.1-2 plants (6–7 weeks old) were infected with Pst DC3000 strains (1 × 107 cfu/ml), or treated with 10 mM MgCl2 as a mock control, and RNA was isolated from leaves for use as RT-PCR template. Primers for the RT-PCR analysis were 90.1a (5′-CAC TAG GGA TGT GGA TGG GGA AC-3′), 90.1b (5′-CAC CTT CGT TTT CTT TCT TTG GTT C-3′) for AtHSP90.1, and actin-F (5′ TCG GTG GTT CCA TTC TTG CT-3′), actin-R (5′-GCT TTT TAA GCC TTT GAT CTT GAG AG-3′) for Arabidopsis actin2 (At3g18780).
Results
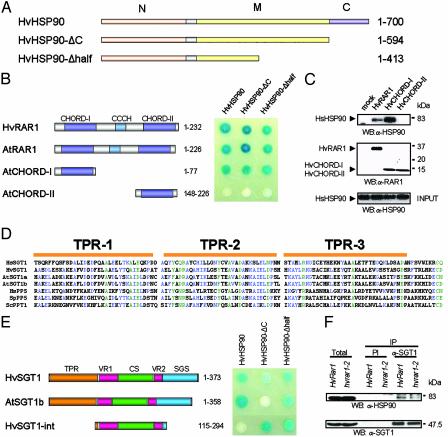
RAR1 Interacts with HSP90. Previous yeast two-hybrid analysis showed that SGT1 binds CHORD-II but not CHORD-I of RAR1, although these domains are highly related (4, 9). To obtain CHORD-I-specific binding proteins, we performed yeast two-hybrid screening by using CHORD-I of barley RAR1 (HvRAR1) as bait. A cytosolic HSP90 (HvHSP90) was identified as an interacting protein (Fig. 1 A and B). The HvHSP90 clone was incomplete at the 3′ end, and we used RT-PCR methods to clone the corresponding full-length cDNA. HvHSP90 shares high identity with human (70%) and Arabidopsis (≈90%) homologs. Both HvRAR1 and Arabidopsis RAR1 (AtRAR1, At5g51700) interacted with HvHSP90, indicating that the HSP90-binding function of plant RAR1 has been conserved across monocots and dicots (Fig. 1B). Consistent with this conservation, in a yeast two-hybrid screen of Arabidopsis using CHORD-I of AtRAR1 as bait, we found Arabidopsis HSP90.1 (AtHSP90.1) as an interacting protein (data not shown). Deletion analysis of AtRAR1 indicated that CHORD-I, but not CHORD-II, interacts with HvHSP90, suggesting that the interaction is highly specific (Fig. 1B).
Fig. 1.
Interaction of HSP90 with RAR1 and SGT1. (A) Domain structures of HvHSP90 constructs. Numbers refer to amino acids encoded. N, N-terminal ATPase domain; M, middle substrate binding domain; C, C-terminal for dimerization and cochaperone binding. (B) In vivo interaction analysis of RAR1 and HSP90 by the yeast two-hybrid system. (Left) Domain structures of plant RAR1 constructs. Interactions were performed by using the LexA system with the lacZ reporter gene, expressing RAR1 proteins as binding domain fusions and HvHSP90 proteins as activator domain fusions. (C) In vitro binding assay using HsHsp90 and S-tag RAR1 fusion derivatives. The precipitated proteins were immunoblotted with the indicated antibodies. Molecular mass markers are indicated (in kDa). (D) Sequence alignment of the TPR domain from SGT1 and PP5 proteins: human SGT1 (Hs, residues 10–123, GenBank accession no. AAD30062), barley SGT1 (Hv, 5–118, AAL33610), Arabidopsis SGT1a and SGT1b (At, 1–114, At4g23570 and At4g11260, respectively), human PP5 (Hs, 27–140, AAD22669), Schizosaccharomyces pombe PP5 (Sp, 4–117, T40391), Saccharomyces cerevisiae PPT1 (Sc, 11–124, S52571). Green indicates 100% conserved residues, and blue indicates >50% conserved residues. (E) In vivo interaction analysis of SGT1 and HSP90 by the yeast two hybrid system. Domain structures of HvSGT1 and AtSGT1b made in pLexA vector are shown on the left. Interactions were detected as shown in B. (F) Coimmunoprecipitation of HvSGT1 and HvHSP90 in barley. Protein extract from HvRAR1 or mutant hvrar1–2 barley plants were immunoprecipiated with SGT1 or preimmune (PI) antibodies. Samples of eluted fractions were analyzed by immunoblotting with antibodies to SGT1 and HSP90.
Cytosolic HSP90 contains three distinct domains: an N-terminal ATPase domain (N), a substrate (or often called “client”) binding domain in the middle (M), and the C-terminal end (C) containing a dimerization domain and a MEEVD motif that binds tetratricopeptide repeat (TPR) domains of many cochaperones (Fig. 1 A) (17, 18). Deletion analysis showed that the N-terminal half of HvHSP90 containing the ATPase domain is sufficient for binding both barley and Arabidopsis RAR1 (Fig. 1B). To test whether RAR1 interacts directly with HSP90, in vitro pull-down experiments were performed. Because purification of E. coli-expressed barley HSP90 proteins was not successful (data not shown), we used HsHsp90 for the analysis. We found that HvRAR1 and HvCHORD-I interact with HsHsp90, whereas HvCHORD-II does not (Fig. 1C). These data indicate that the interaction between RAR1 and HSP90 is direct, specific and highly conserved across eukaryotes.
SGT1 Interacts with HSP90. SGT1 proteins contain a TPR domain closely related to that of protein phosphatase 5 (PP5) that was demonstrated to bind HSP90 (Fig. 1D) (19). This finding prompted us to test whether HvHSP90 also interacts with plant SGT1 proteins in vivo, by using the yeast two-hybrid system. We tested barley SGT1 (HvSGT1) and Arabidopsis SGT1b (AtSGT1b) (9, 10) and found that they both interacted with full-length HvHSP90, indicating that the SGT1-HSP90 interaction is conserved between monocots and dicots (Fig. 1E). The TPR domain of PP5 interacts with the C-terminal end of Hsp90 containing the MEEVD motif (20, 21). We therefore tested whether the C terminus of HvHSP90 is required for SGT1 binding. We found that a C-terminal deletant of HvHSP90 (HvHSP90-ΔC; amino acids 1–594) bound neither HvSGT1 nor AtSGT1b as effectively as full-length HvHSP90 (Fig. 1E). However, a further deletion of HvHSP90 from the C terminus, leaving only the N-terminal half of the protein, restored its ability to bind HvSGT1 and AtSGT1b. These data indicate that HvHSP90 may have two interaction domains for SGT1, one at the C terminus that binds the TPR domain of SGT1 and the other at the N-terminal end, and that the internal region of HvHSP90 between residues 414 and 594 has an inhibitory effect on the binding at the N-terminal end. The CS (CHORD-containing protein and SGT1) domain of SGT1 has been suggested to form a structure similar to p23 that interacts with the ATPase domain of HSP90 (22, 23). Therefore, the CS domain of SGT1 may interact with the ATPase domain of HSP90. To investigate this possibility, we tested the internal region of HvSGT1 containing the CS domain (amino acids 115–294; HvSGT1-int) for interaction with HvHSP90 (Fig. 1E). HvSGT1-int did not interact with full-length HvHSP90, but it did interact with HvHSP90-ΔC and HvHSP90-Δhalf. This observation is consistent with the idea that HvSGT1 and HvHSP90 interact via two distinct domains and the interaction between the CS domain of HvSGT1 and N terminus of HvHSP90 is regulated by other domains. Coimmunoprecipition experiments using anti-HvSGT1 and anti-HvHSP90 antibodies confirmed that HvSGT1 interacts with HvHSP90 in barley (Fig. 1F). The interaction was also detected in rar1-2 mutant plants, indicating that it is HvRAR1 independent.
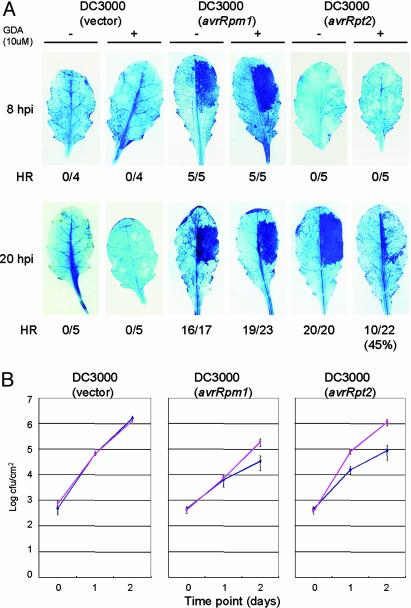
GDA Inhibits RPS2-Dependent HR and Resistance. The specific interaction between HSP90 and both RAR1 and SGT1 prompted us to investigate the involvement of HSP90 in disease resistance. We addressed this question firstly by using GDA. GDA inhibits the ATPase activity of HSP90 by blocking its highly conserved ATP-binding pocket, and has little effect on prokaryotic pathogens (24, 25). In Arabidopsis, GDA can be applied externally and inhibits HSP90 effectively at concentrations >1 μM (the dissociation constant for binding HSP90 is 1.2 μM) (26). We tested whether GDA affects the HR triggered by the Arabidopsis R genes RPM1 and RPS2 upon specific recognition of Pseudomonas syringae pv. tomato (Pst) strains containing the corresponding avirulence genes avrRpm1 and avrRpt2. By trypan blue staining, the RPM1- and RPS2-dependent HRs are normally visible 5–8 h and 15–20 h postinoculation (hpi), respectively (27, 28). Application of 10 μM GDA did not noticeably affect the RPM1-dependent HR at 8 hpi (Fig. 2A). Higher concentrations of GDA (50 μM) also failed to inhibit the RPM1-dependent HR at 8 hpi (data not shown). In contrast, GDA at 10 μM severely diminished the RPS2-dependent HR at 20 hpi. Only 45% of leaves that were inoculated with Pst DC3000 (avrRpt2) exhibited the HR phenotype at 20 hpi, whereas other leaves showed cell death only at the edge of the inoculated area. Because both AvrRpt2 and AvrRpm1 are type III effectors and the RPM1-dependent HR was not affected by GDA, it is unlikely that GDA inhibits the type III delivery system in Pst DC3000. These data suggest that HSP90 is important for RPS2-dependent HR cell death.
Fig. 2.
Geldanamycin inhibits RPS2-dependent HR and resistance. (A) The HR test by trypan blue staining. The right half of the leaves from 6-week-old Arabidopsis Col-0 plants (containing both RPM1 and RPS2) were infiltrated with Pst DC3000 strains (1 × 107 cfu/ml) containing vector only, or clones expressing avrRpm1 or avrRpt2. GDA (10μM) or mock solutions lacking GDA were infiltrated together with the bacterial pathogens. Leaves were stained with trypan blue 8 or 20 hpi. Fractions indicate numbers of leaves exhibiting HR and total number of leaves tested. (B) Bacterial growth analysis of Pst DC3000 strains (1 × 105 cfu/ml) containing vector only, or clones expressing avrRpm1 or avrRpt2, inoculated into Arabidopsis Col-0, together with GDA (pink line) or solution lacking GDA (blue line). These experiments were performed three times with similar results.
We also tested whether GDA affects the R-gene mediated resistance against Pst DC3000 (avrRpm1 or avrRpt2) strains (Fig. 2B). For Pst DC3000 (avrRpm1), GDA had no significant effect on bacterial growth by 1 day postinoculation (dpi), but by 2 dpi, GDA enabled an slight increase in bacterial titer (≈6-fold). GDA showed a more pronounced inhibition of RPS2 resistance against Pst DC3000 (avrRpt2), resulting in ≈5- to 6-fold higher titer by 1 dpi, and >10-fold higher titer by 2 dpi. GDA did not affect growth of virulent Pst DC3000 (vector). Taken together, these data indicate that HSP90 activity is required for both full RPS2 and RPM1-dependent disease resistance, in addition to the RPS2 triggered HR.
AtHSP90.1 Is Induced by Pst DC3000. The Arabidopsis genome contains four genes for cytosolic HSP90 (previously named HSP82), AtHSP90.1 (At5g52640), AtHSP90.2 (At5g56030), AtHSP90.3 (At5g56010), and AtHSP90.4 (At5g56000) (29, 30). It should be noted that most eukaryotes contain more than one cytosolic HSP90 isoform, and that there is little evidence so far that these genes have specialized functions (24). Although Arabidopsis HSP90 isoforms are highly related (>85% identical), they show distinct expression profiles (29). For example, AtHSP90.1 is expressed significantly only after heat shock, whereas AtHSP90.2 and AtHSP90.3 are constitutively expressed and only moderately induced by heat treatment (29, 31). AtHSP90.2, AtHSP90.3, and AtHSP90.4 encode proteins that are 97% identical and are located next to each other on Arabidopsis chromosome 5, suggesting that they are the result of recent gene duplication.
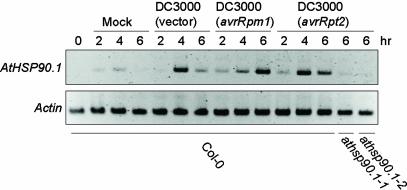
We tested by RT-PCR the expression of the AtHSP90 isoforms after infection with various Pst DC3000 strains. AtHSP90.1 expression was not detectable before inoculation, consistent with previous reports of low or no AtHSP90.1 expression in the absence of stress (Fig. 3) (29, 31). However, upon inoculation with virulent Pst DC3000 (vector), AtHSP90.1 expression was detectable by 4 hpi, but was reduced at 6 hpi. AtHSP90.1 expression was similar in response to inoculation with Pst DC3000 (avrRpt2). The expression level of AtHSP90.1 after treatment with Pst DC3000 (avrRpm1) was much higher at 6 hpi than at 4 hpi. This strong expression of AtHSP90.1 at 6 hpi may partly reflect the earlier onset of HR triggered by RPM1, at around 5hpi. Mock inoculation provided an abiotic stimulus resulting in weak AtHSP90.1 induction. We did not detect significant induction of other HSP90 isoforms (data not shown), suggesting that AtHSP90.1 is the only cytosolic HSP90 in Arabidopsis to be induced significantly by Pst DC3000.
Fig. 3.
AtHSP90.1 mRNA expression is induced by Pst DC3000 strains. RT-PCR analysis was performed on mRNA isolated from leaves inoculated with Pst DC3000 strains (1 × 107 cfu/ml) containing vector only, or clones expressing avrRpm1 or avrRpt2 or leaves subjected to mock inoculation. RT-PCR from an actin gene was used as a control to verify evenness of RNA template amounts.
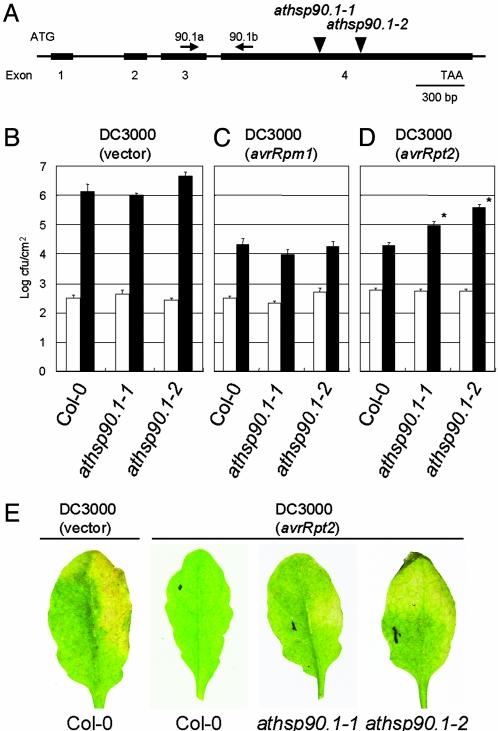
The AtHSP90.1 Isoform Is Required for Full RPS2-Mediated Resistance. The pathogen inducibility of AtHSP90.1 prompted us to test whether it is required for disease resistance. We obtained two distinct T-DNA insertion lines of AtHSP90.1 (see Materials and Methods) in Col-0 background and selected homozygous mutant plants by PCR. We refer to these mutants as athsp90.1-1 (Salk_007614) and athsp90.1-2 (Salk_075596) (Fig. 4A). RT-PCR using AtHSP90.1-specific primers on pathogen inoculated mutant plants gave no detectable amplification products, indicating that AtHSP90.1 transcript was absent or severely reduced (Fig. 3). Mutant athsp90.1-1 and athsp90.1-2 plants showed no obvious morphological defects. Growth of virulent Pst DC3000 (vector) was not affected by the athsp90.1 mutations, suggesting that AtHSP90.1-1 is not required for “basal defence” (32) (Fig. 4B). The mutants also did not show loss of RPM1-dependent resistance (Fig. 4C). However, both athsp90.1-1 and athsp90.1-2 mutations compromised RPS2-dependent resistance, resulting in 5- to 20-fold more growth by 3 dpi, and increased disease chlorosis by 6 dpi (Fig. 4 D and E). Pst DC3000 (avrRpt2) did not grow in the athsp90.1 mutants to the same extent as in Pst DC3000 (vector), indicating a partial loss of RPS2 resistance. It is plausible that the other isoforms of HSP90 partially compensated for the loss of AtHSP90.1. The athsp90.1-1 mutation did not noticeably affect growth of Peronospora parasitica isolates that were virulent (Noco2), or avirulent (Cala2), with respect to the wild-type Col-0 plants (data not shown). In summary, the genetic analysis demonstrated a requirement for HSP90 in disease resistance, although the known range of this effect is so far limited to the requirement for AtHSP90.1 in full RPS2-mediated resistance.
Fig. 4.
AtHSP90.1 is required for RPS2-dependent resistance. (A) Relative position of T-DNA insertions within the AtHSP90.1 gene. Exons are indicated by black boxes. Primers used for RT-PCR were indicated. (B) Bacterial growth analysis of Pst DC3000 (vector) (1 × 105 cfu/ml), which was hand-infiltrated with the needleless syringe into the leaves of 6- to 7-week-old Arabidopsis Col-0, or mutants athsp90.1-1 and athsp90.1-2. Leaves were harvested immediately after infiltration (white column) or at 3 dpi (black column). (C) Same as in B, except with Pst DC3000 (avrRPM1). (D) Same as in B, except with Pst DC3000 (avrRpt2). *, Significantly different from wild-type control at P < 0.05. (E) Disease phenotype of athsp90.1-1 and athsp90.1-2. The right halves of leaves were hand-infiltrated with Pst DC3000 (vector) or Pst DC3000 (avrRpt2) (1 × 105 cfu/ml), and photographs were taken 6 dpi.
Discussion
HSP90 Is Essential for RPS2-Dependent Resistance. We and others recently demonstrated that RAR1 and its interacting protein SGT1 are important components of plant disease resistance triggered by a number of R proteins (33). In this report, we demonstrate that RAR1 and SGT1 specifically interact with HSP90, and that HSP90 activity is required for RPS2 resistance, and at least partially required for RPM1 resistance. Full expression of both RPS2 and RPM1 resistance in Arabidopsis has been found to require RAR1 (5, 6) but not AtSGT1b (10, 13). However, it is possible that the second Arabidopsis SGT1 isoform, AtSGT1a, can compensate for the loss of AtSGT1b. Consistent with this idea, virus-induced gene silencing of Nb-SGT1 in N. benthamiana, which presumably targeted all SGT1 homologs, compromised the HR induced by AvrRpt2 expression (11). Taken together, our data demonstrate a requirement for HSP90 in R protein-mediated disease resistance, and furthermore, that this activity may occur via direct interactions with RAR1 and SGT1.
The apparent specific requirement for AtHSP90.1 in RPS2- but not RPM1-dependent resistance, and the fact that GDA inhibited RPS2 resistance to a greater degree than RPM1 resistance, is intriguing. One hypothesis is that the RPM1 signaling pathway may be partially independent of HSP90 activity per se. Another possibility is that RPM1 and RPS2 may have differential requirements for the different HSP90 isoforms. Mutations in a constitutively expressed HSP90 isoform, AtHSP90.2, abolish RPM1 resistance, supporting such idea (J. Dangl, personal communication). The HR triggered by RPM1 occurs earlier than that triggered by RPS2 (5 h versus 15 h), suggesting that RPM1 activates signaling earlier than RPS2. It may be that RPM1 signaling began before GDA had effectively bound all HSP90 molecules.
RAR1 and SGT1 May Function As Cochaperones of HSP90. HSP90 functions in protein complexes with a large set of cochaperones (24). Cochaperones can be classified according to whether or not they contain TPR domains. The TPR-type cochaperones such as Hop (Sti1 in yeast), PP5, and immunophilins, physically associate with the C-terminal MEEVD motif of Hsp90. Non-TPR-type cochaperones such as Cdc37 and p23 (Sba1 in yeast) also interact with Hsp90 but in a MEEVD independent manner. RAR1 that does not possess a TPR domain interacts with the N-terminal half of HSP90. The TPR domain of SGT1 is highly related to those of Hop and PP5 which interact with the C-terminal domain of HSP90. We further demonstrated that the internal CS region of SGT1 provides an additional HSP90-binding site that can associate with the N terminus of HSP90. Based on their structure and interaction partners, SGT1 and RAR1 may be classified as TPR- and non-TPR-type cochaperones, respectively. Our data indicate that the TPR type cochaperone SGT1 interacts with non-TPR type cochaperone RAR1. Interestingly, an interaction between TPR and non-TPR cochaperones has been reported (34). The TPR type cochaperone Hop/Sti1 physically and genetically interacts with the non-TPR type cochaperone cdc37, providing a multiplicity of chaperone complex formation (34, 35). How HSP90 interacts specifically with its substrate proteins and how HSP90 activity is regulated is unknown, although cochaperones, possibly including RAR1 and SGT1, are thought to play an important role in these processes.
Potential Substrates of HSP90 in Disease Resistance Signaling. Recent studies of animal and yeast Hsp90 indicate that substrates of Hsp90 chaperones are principally factors involved in signaling, such as steroid receptors/transcription factors and protein kinases (24). Therefore it is possible that the HSP90 chaperone complex with RAR1 and SGT1 modulates activity and/or stability of substrate proteins that are essential for disease resistance signaling. The hallmark characteristic of HSP90 substrates is that they become unstable when HSP90 activity is inhibited (24). In light of this, R proteins are candidate HSP90 substrates, because silencing of HSP90 lead to a reduction in the abundance of the R protein Rx that confers resistance to potato virus X (D. Baulcombe, personal communication). Significantly, RPM1 is unstable in rar1 (5) and athsp90.2 mutants (J. Dangl, personal communication), consistent with the idea that R proteins are HSP90 substrates and RAR1 may be a cochaperone of HSP90. This model is also consistent with the recent finding that RAR1 dependency is conditioned by subtle intrinsic properties of barley R proteins MLA1 and MLA6 (36). MLA1 does not require RAR1, whereas MLA6 does, although these proteins are >91% identical (37). In this model, MLA6, but not MLA1, may require RAR1 to achieve proper conformation and stability.
Several pieces of evidence suggest that SGT1 plays diverse roles in plants, whereas RAR1 plays more specialized resistance functions (33). Only a subset of R proteins require RAR1, whereas SGT1 is essential for disease resistance and HR triggered by a wide range of R proteins including non-leucine-rich repeats (LRR) type R proteins (11). AtRAR1 is not essential for viability (5, 6), whereas the atsgt1a-1/atsgt1b-1 double mutant combination is embryo lethal (A.T. and K.S. unpublished data). Furthermore, AtRAR1 is not required for the correct auxin response controlled by an SCF complex, whereas AtSGT1b is essential for this response (14). SGT1, RAR1, and HSP90 may also have common and distinct function specifically associated with disease resistance. For example, combining atrar1-10 and atsgt1b–1 mutations has an additive effect on resistance, suggesting that RAR1 and SGT1b can act at least partially independently of one another (10). Furthermore, the Rx protein shows reduced abundance in plants silenced for HSP90 but not SGT1 (38). One speculative model to explain the disparate function of RAR1, SGT1, and HSP90 in resistance is that HSP90 stabilizes R proteins, in some cases together with RAR1 (e.g., for RPM1), and SGT1 is then recruited to modulate activity of R proteins. Because yeast Sgt1 interacts with various proteins containing LRR (22), it will be of particular interest to see whether plant SGT1 and HSP90 associate with the LRRs of R proteins.
HSP90 and SGT1 may also function downstream of R protein signaling pathways. HSP90 is known to associate with large protein complexes including the proteasome, SCF complex and the components of the centromere-binding factor 3 (CBF3) kinetochore (39–41). Biochemical functions of HSP90 in these complexes are still unknown. One possible scenario is that formation or activation of such protein complexes requires the chaperoning activity of HSP90. In yeast, for example, formation of the CBF3 kinetochore complex requires activation of Ctf13 by Hsp90 (41) and interaction of Sgt1 with Skp1 and Hsp90 (ref. 8 and K. Kitagawa, personal communication). Because Ctf13 is an F-box protein, it is also possible that Hsp90, together with Sgt1, can activate other Skp1-F-box protein complexes such as the SCF complex. Consistent with this idea, both mouse Sgt1 and Hsp90 are found in the SCFSKP2 complex that meditates ubiquitylation of the cyclin-dependent kinase inhibitor p27 (40). By analogy, plant SGT1 and HSP90 may regulate the activity of SCF complexes that mediate degradation or activation of regulators of disease resistance.
RAR1 Homologs in Animals. The structure of SGT1 and HSP90 is highly conserved and their functions are essential for viability in yeast and plants. On the other hand, yeast lack a RAR1 homolog, and rar1 mutant plants have no visible growth defects, indicating that RAR1 is not an essential component for cell viability in eukaryotes per se. Interestingly, metazoan RAR1 homologs, termed CHORD-containing proteins (Chp), possess a C-terminal domain related to the CS motif of SGT1 that interacts with HSP90 (4). Furthermore, plant RAR1 interacts with HsHsp90, implying that the biochemical function of RAR1 may be at least in part conserved in these organisms. In this context, it is particularly interesting that Melusin, the animal RAR1 homolog, is involved in stress signaling in the heart (7). Thus, it will be interesting to know whether Melusin mediates the stress signal via Hsp90 and Sgt1. Clearly, further biochemical characterization is needed to understand how RAR1, HSP90, and SGT1 affect signaling processes that may be shared by both animals and plants.
Acknowledgments
We thank Pietro Piffanelli and Paul Schulze-Lefert for the barley cDNA library used in the yeast two-hybrid screening; Lina Kvant and Cristina Azevedo for technical assistance; Katsumi Kitagawa, Jeff Dangl, and David Baulcombe for communicating unpublished results; and Jack Peart and Nick Collins for critical reading of the manuscript. We also thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation. This work was supported by The Gatsby Charitable Foundation (K.S.), the U.K. Biotechnology and Biological Sciences Research Council (K.S.), and the Japan Society for the Promotion of Sciences (K.I.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GDA, geldanamycin; HR, hypersensitive response; TPR, tetratricopeptide repeat; CHORD, cysteine- and histidine-rich domain; SCF, Skp1–Cul1–F box; R, resistance; Hsp, heat shock protein; dpi, days postinoculation; hpi, hours postinoculation; cfu, colony-forming unit.
References
- 1.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- 2.Shirasu, K. & Schulze-Lefert, P. (2000) Plant Mol. Biol. 44, 371–385. [DOI] [PubMed] [Google Scholar]
- 3.Staskawicz, B. J., Mudgett, M. B., Dangl, J. L. & Galan, J. E. (2001) Science 292, 2285–2289. [DOI] [PubMed] [Google Scholar]
- 4.Shirasu, K., Lahaye, L., Tan, M.-W., Zhou, F., Azevedo, C. & Schulze-Lefert, P. (1999) Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- 5.Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R. W. & Dangl, J. L. (2002) Plant Cell 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muskett, P. R., Kahn, K., Austin, M. J., Moisan, L. J., Sadanandom, A., Shirasu, K., Jones, J. D. G. & Parker, J. E. (2002) Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brancaccio, M., Fratta, L., Notte, A., Hirsch, E., Poulet, R., Guazzone, S., De Acetis, M., Vecchione, C., Marino, G., Altruda, F., et al. (2003) Nat. Med. 9, 68–75. [DOI] [PubMed] [Google Scholar]
- 8.Kitagawa, K., Skowyra, D., Elledge, S. J., Harper, J. W. & Hieter, P. (1999) Mol. Cell 4, 21–33. [DOI] [PubMed] [Google Scholar]
- 9.Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K. & Schulze-Lefert, P. (2002) Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- 10.Austin, M. J., Muskett, P., Kahn, K., Feys, B. J., Jones, J. D. & Parker, J. E. (2002) Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- 11.Peart, J. R., Lui, R., Sadanandom, A., Malcuit, I., Moffett, P., Brice, D. C., Schauser, L., Jaggard, D. A. W., Xiao, S., Coleman, M. J., et al. (2002) Proc. Acad. Natl. Sci. USA 99, 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, Y., Schiff, M., Serino, G., Deng, X. W. & Dinesh-Kumar, S. P. (2002) Plant Cell 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tör, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J. L. & Holub, E. B. (2002) Plant Cell 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray, W. M., Muskett, P. R., Chuang, H.-W. & Parker, J. E. (2003) Plant Cell 15, 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katagiri, F., Thilmony, R. & He, S. Y. (2002) in The Arabidopsis Book, eds. Somerville, C. & Meyerowitz, E. (Am. Soc. Plant Biol., Rockville, MD), doi/10.1199/tab.0039.
- 16.Koch, E. & Slusarenko, A. (1990) Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, P., Prodromou, C., Hu, B., Vaughan, C., Roe, S. M., Panaretou, B., Piper, P. W. & Pearl, L. H. (2003) Mol. Cell 11, 647–658. [DOI] [PubMed] [Google Scholar]
- 18.Pearl, L. H. & Prodromou, C. (2000) Curr. Opin. Struct. Biol. 10, 46–51. [DOI] [PubMed] [Google Scholar]
- 19.Chinkers, M. (2001) Trends Endocrinol. Metab. 12, 28–32. [DOI] [PubMed] [Google Scholar]
- 20.Russell, L. C., Whitt, S. R., Chen, M. S. & Chinkers, M. (1999) J. Biol. Chem. 274, 20060–20063. [DOI] [PubMed] [Google Scholar]
- 21.Scheufler, C., Brinker, A., Bourenkov, G., Pegoraro, S., Moroder, L., Bartunik, H., Hartl, F. U. & Moarefi, I. (2000) Cell 101, 199–210. [DOI] [PubMed] [Google Scholar]
- 22.Dubacq, C., Guerois, R., Courbeyrette, R., Kitagawa, K. & Mann, C. (2002) Eukaryot. Cell 1, 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Ranea, J. A., Mirey, G., Camonis, J. & Valencia, A. (2002) FEBS Lett. 529, 162–167. [DOI] [PubMed] [Google Scholar]
- 24.Picard, D. (2002) Cell. Mol. Life Sci. 59, 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stebbins, C. E., Russo, A. A., Schneider, C., Rosen, N., Hartl, F. U. & Pavletich, N. P. (1997) Cell 89, 239–250. [DOI] [PubMed] [Google Scholar]
- 26.Queitsch, C., Sangster, T. A. & Lindquist, S. (2002) Nature 417, 618–624. [DOI] [PubMed] [Google Scholar]
- 27.Axtell, M. J. & Staskawicz, B. J. (2003) Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- 28.Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R. & Dangl, J. L. (2003) Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- 29.Yabe, N., Takahashi, T. & Komeda, Y. (1994) Plant Cell Physiol. 35, 1207–1219. [DOI] [PubMed] [Google Scholar]
- 30.Krishna, P. & Gloor, G. (2001) Cell Stress Chaperones 6, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haralampidis, K., Milioni, D., Rigas, S. & Hatzopoulos, P. (2002) Plant Physiol. 129, 1138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond-Kosack, K. E. & Parker, J. (2003) Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- 33.Shirasu, K. & Schulze-Lefert, P. (2003) Trends Plant Sci. 8, 252–258. [DOI] [PubMed] [Google Scholar]
- 34.Abbas-Terki, T., Briand, P. A., Donze, O. & Picard, D. (2002) Biol. Chem. 383, 1335–1342. [DOI] [PubMed] [Google Scholar]
- 35.Hartson, S. D., Irwin, A. D., Shao, J. Y., Scroggins, B. T., Volk, L., Huang, W. J. & Matts, R. L. (2000) Biochemistry 39, 7631–7644. [DOI] [PubMed] [Google Scholar]
- 36.Shen, Q., Zhou, F., Bieri, S., Haizel, H., Shirasu, K. & Schulze-Lefert, P. (2003) Plant Cell 15, 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, F., Kurth, J. C., Wei, F., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R. & Schulze-Lefert, P. (2001) Plant Cell 13, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffett, P., Farnham, G., Peart, J. & Baulcombe, D. C. (2002) EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eleuteri, A. M., Cuccioloni, M., Bellesi, J., Lupidi, G., Fioretti, E. & Angeletti, M. (2002) Proteins 48, 169–177. [DOI] [PubMed] [Google Scholar]
- 40.Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C. S., Wolf, D. A., Wei, N. & Deshaies, R. J. (2001) Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- 41.Stemmann, O., Neidig, A., Kocher, T., Wilm, M. & Lechner, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8585–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]