Abstract
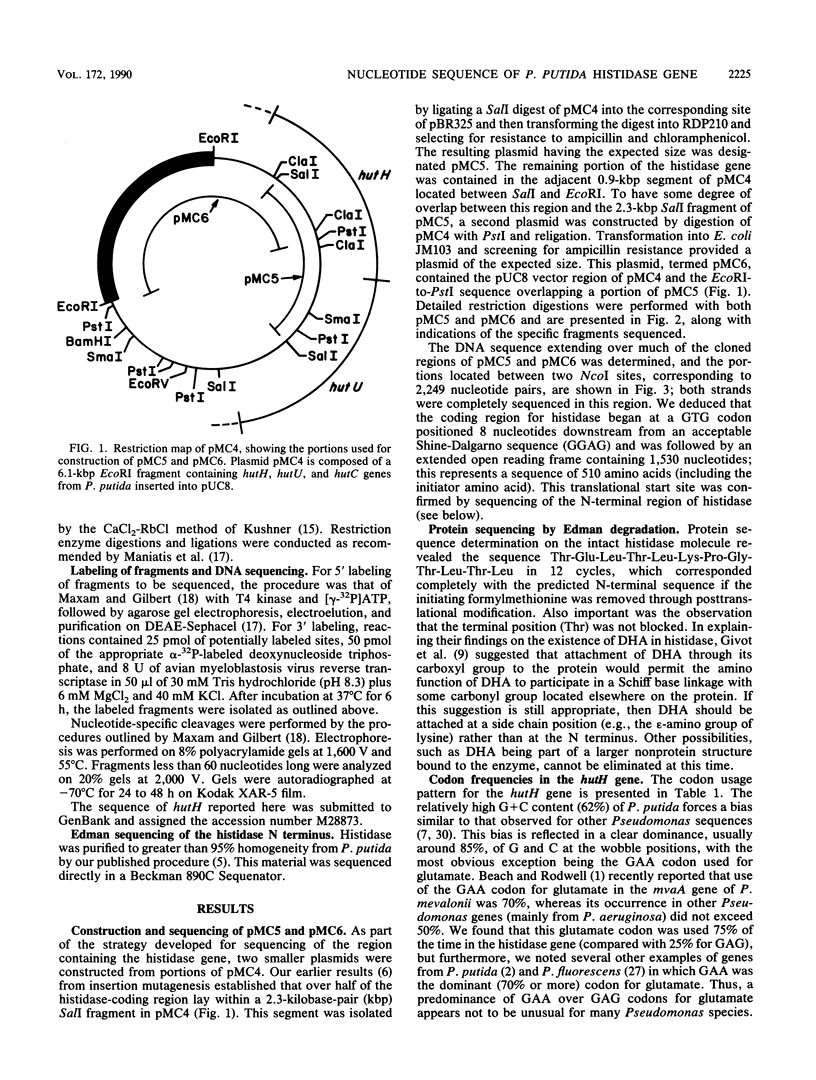
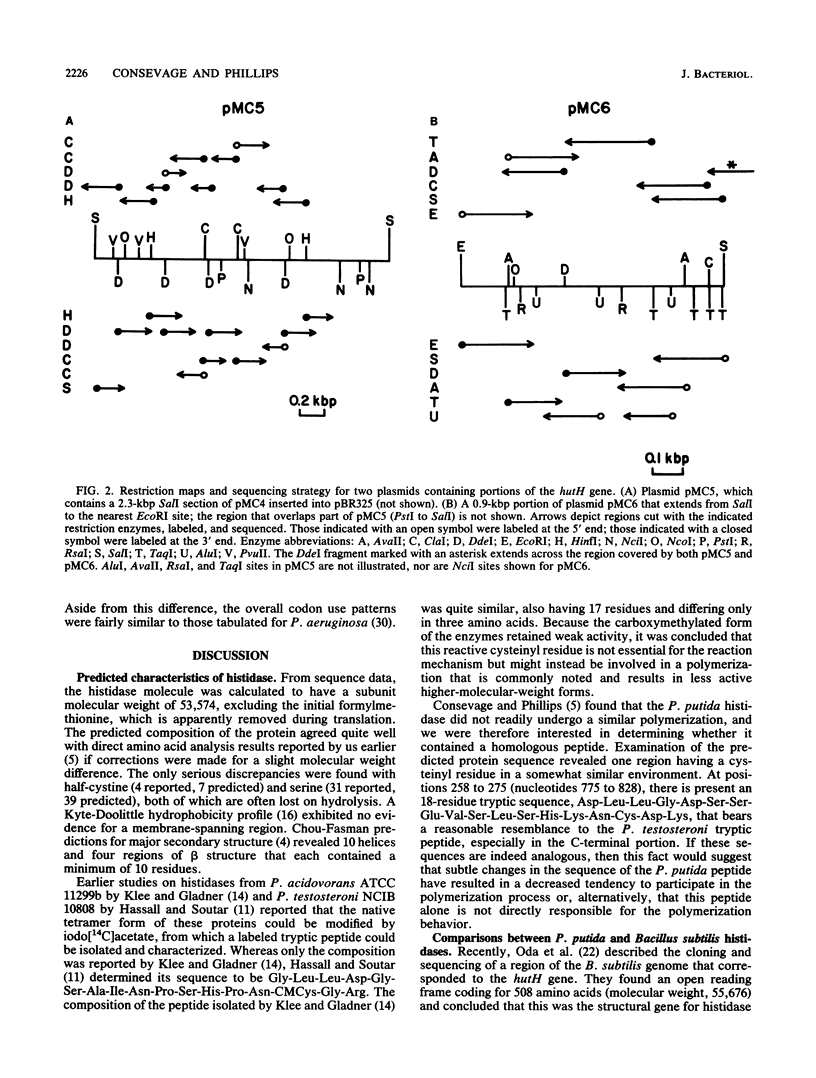
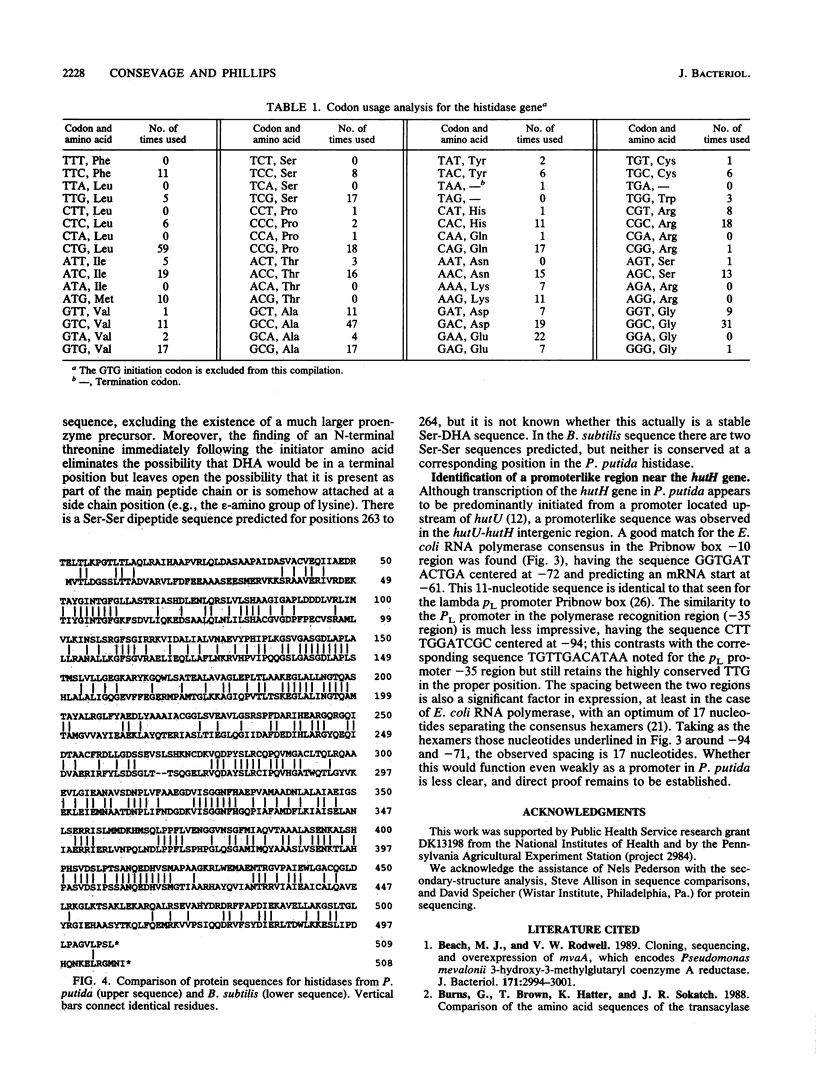
The complete nucleotide sequence of the hutH gene, encoding histidine ammonia-lyase (histidase), in Pseudomonas putida ATCC 12633 has been determined from the appropriate portions of the hut region that had been cloned into Escherichia coli. The resulting DNA sequence revealed an open reading frame of 1,530 base pairs, corresponding to a protein subunit of approximate molecular weight 53,600, in the location previously identified for the histidase gene by Tn1000 mutagenesis. Translation began at a GTG codon, but direct protein sequencing revealed that the initiating amino acid was removed posttranslationally to provide an N-terminal threonine; 11 additional residues completely agreed with the predicted amino acid sequence. This sequence excluded the possibility that a dehydroalanine unit, the postulated coenzyme for histidase, is attached at the N terminus of histidase subunits. Comparison of the P. putida histidase gene sequence with that of a Bacillus subtilis region encoding histidase revealed 42% identity at the protein level. Although the hutU (urocanase) and hutH (histidase) genes are induced by urocanate and normally are transcribed as a unit beginning with hutU, analysis of the region immediately upstream of the histidase gene revealed a potential weak promoter that may possibly be used to maintain a basal level of histidase for the generation of inducer (urocanate) when histidine levels are elevated.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beach M. J., Rodwell V. W. Cloning, sequencing, and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Bacteriol. 1989 Jun;171(6):2994–3001. doi: 10.1128/jb.171.6.2994-3001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns G., Brown T., Hatter K., Sokatch J. R. Comparison of the amino acid sequences of the transacylase components of branched chain oxoacid dehydrogenase of Pseudomonas putida, and the pyruvate and 2-oxoglutarate dehydrogenases of Escherichia coli. Eur J Biochem. 1988 Sep 1;176(1):165–169. doi: 10.1111/j.1432-1033.1988.tb14264.x. [DOI] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Consevage M. W., Phillips A. T. Presence and quantity of dehydroalanine in histidine ammonia-lyase from Pseudomonas putida. Biochemistry. 1985 Jan 15;24(2):301–308. doi: 10.1021/bi00323a010. [DOI] [PubMed] [Google Scholar]
- Consevage M. W., Porter R. D., Phillips A. T. Cloning and expression in Escherichia coli of histidine utilization genes from Pseudomonas putida. J Bacteriol. 1985 Apr;162(1):138–146. doi: 10.1128/jb.162.1.138-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford I. P., Eberly L. Structure and regulation of the anthranilate synthase genes in Pseudomonas aeruginosa: I. Sequence of trpG encoding the glutamine amidotransferase subunit. Mol Biol Evol. 1986 Sep;3(5):436–448. doi: 10.1093/oxfordjournals.molbev.a040408. [DOI] [PubMed] [Google Scholar]
- Givot I. L., Abeles R. H. Mammalian histidine ammonia lyase. In vivo inactivation and presence of an electrophilic center at the active site. J Biol Chem. 1970 Jun;245(12):3271–3273. [PubMed] [Google Scholar]
- Givot I. L., Smith T. A., Abeles R. H. Studies on the mechanism of action and the structure of the electrophilic center of histidine ammonia lyase. J Biol Chem. 1969 Dec 10;244(23):6341–6353. [PubMed] [Google Scholar]
- Hadero A., Crawford I. P. Nucleotide sequence of the genes for tryptophan synthase in Pseudomonas aeruginosa. Mol Biol Evol. 1986 May;3(3):191–204. doi: 10.1093/oxfordjournals.molbev.a040388. [DOI] [PubMed] [Google Scholar]
- Hassall H., Soutar A. K. Amino acid sequence of a peptide containing the active cysteine residue of histidine ammonia-lyase. Biochem J. 1974 Mar;137(3):559–566. doi: 10.1042/bj1370559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Phillips A. T. Organization and multiple regulation of histidine utilization genes in Pseudomonas putida. J Bacteriol. 1988 Sep;170(9):4272–4279. doi: 10.1128/jb.170.9.4272-4279.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee C. B., Gladner J. A. Isolation of a cysteine-peptide at the active site of histidine ammonia-lyase. J Biol Chem. 1972 Dec 25;247(24):8051–8057. [PubMed] [Google Scholar]
- Klee C. B. Metal activation of histidine ammonia-lyase. Metal ion-sulfhydryl group relationship. J Biol Chem. 1972 Mar 10;247(5):1398–1406. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Brosius J., McClure W. R. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985 Mar 25;260(6):3529–3538. [PubMed] [Google Scholar]
- Oda M., Sugishita A., Furukawa K. Cloning and nucleotide sequences of histidase and regulatory genes in the Bacillus subtilis hut operon and positive regulation of the operon. J Bacteriol. 1988 Jul;170(7):3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Recsei P. A., Snell E. E. Pyruvoyl enzymes. Annu Rev Biochem. 1984;53:357–387. doi: 10.1146/annurev.bi.53.070184.002041. [DOI] [PubMed] [Google Scholar]
- Rokosu A. A. Immunological relatedness of histidine ammonia-lyases from some species of Pseudomonas: taxonomic implication. Int J Biochem. 1983;15(6):867–870. doi: 10.1016/0020-711x(83)90160-x. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Takagi J. S., Tokushige M., Shimura Y. Cloning and nucleotide sequence of the aspartase gene of Pseudomonas fluorescens. J Biochem. 1986 Sep;100(3):697–705. doi: 10.1093/oxfordjournals.jbchem.a121762. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Hermodson M. A., Zalkin H. Primary structure of Serratia marcescens anthranilate synthase component II. J Biol Chem. 1980 Feb 25;255(4):1451–1457. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Dehydroalanine in histidine ammonia lyase. J Biol Chem. 1969 Dec 10;244(23):6550–6552. [PubMed] [Google Scholar]


