Abstract
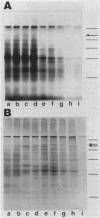
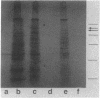
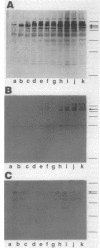
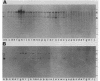
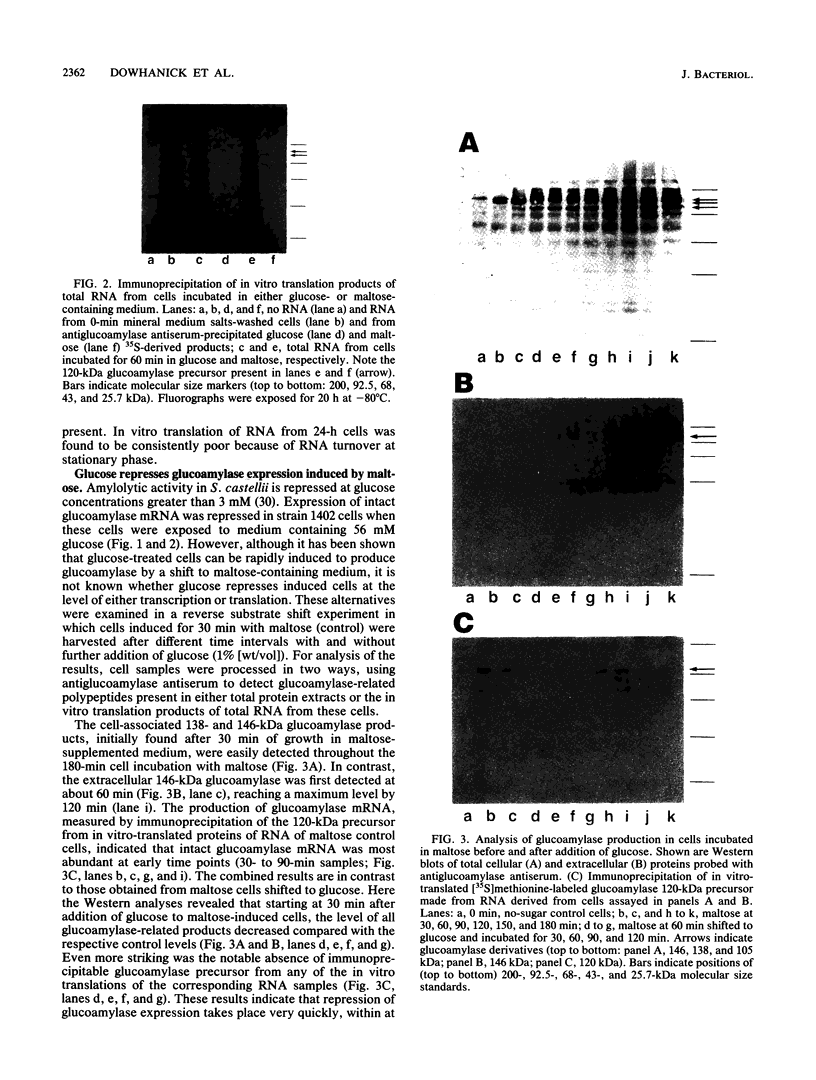
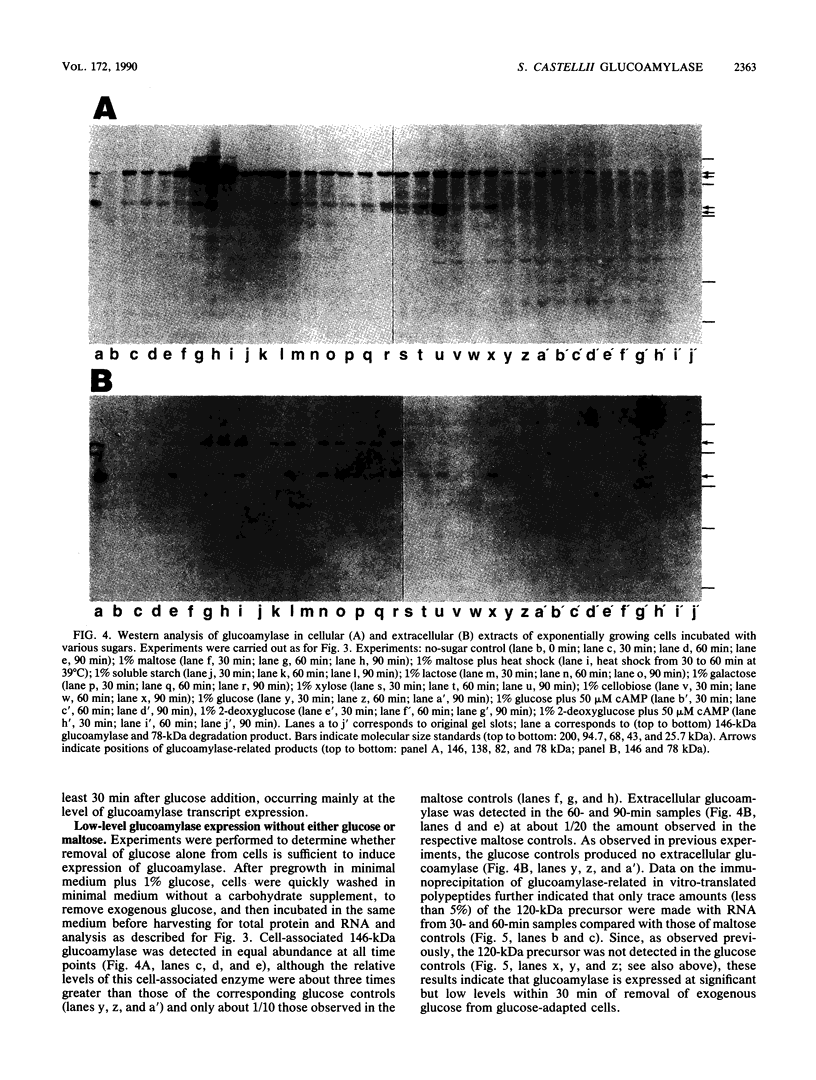
Expression of the 146-kilodalton (kDa) extracellular glucoamylase by the budding yeast Schwanniomyces castellii is induced by maltose and starch. By use of antiglucoamylase antisera, we found that this expression was regulated at the level of the mRNA, taking place within 30 min after exposure of yeast cells to the respective sugars. Polyacrylamide gel electrophoresis analysis of the in vitro-translated products of total RNA from maltose-treated cells established that the glucoamylase precursor was approximately 120 kDa in size. Stable glucoamylase transcript was not produced in cells exposed to glucose, 2-deoxyglucose, and heat shock. Cells exposed to these two sugars also degraded intracellular and extracellular glucoamylase. In the presence of sugars such as cellobiose, galactose, lactose, and xylose or in the absence of any carbohydrate, a low-level, constitutive-like expression of this preglucoamylase occurred. The nascent glucoamylase underwent at least two posttranslational modifications, resulting in a 138-kDa cell-associated form and the 146-kDa active form that was found free in the medium. These results suggest that glucoamylase expression is tightly regulated similarly to expression of the enzymes responsible for maltose metabolism in Saccharomyces yeasts.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boel E., Hjort I., Svensson B., Norris F., Norris K. E., Fiil N. P. Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNAs. EMBO J. 1984 May;3(5):1097–1102. doi: 10.1002/j.1460-2075.1984.tb01935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Clementi F., Rossi J., Costamagna L., Rosi J. Production of amylase(s) by Schwanniomyces castellii and Endomycopsis fibuligera. Antonie Van Leeuwenhoek. 1980;46(4):399–405. doi: 10.1007/BF00421986. [DOI] [PubMed] [Google Scholar]
- Erratt J. A., Douglas P. E., Moranelli F., Seligy V. L. The induction of alpha-amylase by starch in Aspergillus oryzae: evidence for controlled mRNA expression. Can J Biochem Cell Biol. 1984 Aug;62(8):678–690. doi: 10.1139/o84-089. [DOI] [PubMed] [Google Scholar]
- Erratt J. A., Nasim A. Cloning and expression of a Saccharomyces diastaticus glucoamylase gene in Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Bacteriol. 1986 May;166(2):484–490. doi: 10.1128/jb.166.2.484-490.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gines M. J., Dove M. J., Seligy V. L. Aspergillus oryzae has two nearly identical Taka-amylase genes, each containing eight introns. Gene. 1989 Jun 30;79(1):107–117. doi: 10.1016/0378-1119(89)90096-6. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Craig E. A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Mackay R. M., Baird S., Dove M. J., Erratt J. A., Gines M., Moranelli F., Nasim A., Willick G. E., Yaguchi M., Seligy V. L. Glucanase gene diversity in prokaryotic and eukaryotic organisms. Biosystems. 1985;18(3-4):279–292. doi: 10.1016/0303-2647(85)90028-0. [DOI] [PubMed] [Google Scholar]
- Manjunath P., Shenoy B. C., Raghavendra Rao M. R. Fungal glucoamylases. J Appl Biochem. 1983 Aug-Oct;5(4-5):235–260. [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Genetic analysis of the role of cAMP in yeast. Yeast. 1985 Sep;1(1):15–24. doi: 10.1002/yea.320010103. [DOI] [PubMed] [Google Scholar]
- McCann A. K., Barnett J. A. The utilization of starch by yeasts. Yeast. 1986 Jun;2(2):109–115. doi: 10.1002/yea.320020206. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Meade J. H., Cole G., Lawyer F. C., McCabe P., Schweickart V., Tal R., Wittman V. P., Flatgaard J. E., Innis M. A. Molecular cloning and characterization of the glucoamylase gene of Aspergillus awamori. Mol Cell Biol. 1984 Nov;4(11):2306–2315. doi: 10.1128/mcb.4.11.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteng-Gyang K., Moulin G., Galzy P. A study of amylolytic system of Schwanniomyces castelii. Z Allg Mikrobiol. 1981;21(7):537–544. doi: 10.1002/jobm.3630210707. [DOI] [PubMed] [Google Scholar]
- Prakash K., Seligy V. L. A temporally expressed gene from Schwanniomyces alluvius and detection of homologous sequences in other yeasts. Gene. 1988 Dec 15;73(1):131–140. doi: 10.1016/0378-1119(88)90319-8. [DOI] [PubMed] [Google Scholar]
- Pretorius I. S., Modena D., Vanoni M., Englard S., Marmur J. Transcriptional control of glucoamylase synthesis in vegetatively growing and sporulating Saccharomyces species. Mol Cell Biol. 1986 Sep;6(9):3034–3041. doi: 10.1128/mcb.6.9.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlanderer G., Dellweg H. Cyclid AMP and catabolite repression in yeasts, In Schizosaccharomyces pombe glucose lowers both intracellular adenosine 3':5'-monophosphate levels and the activity of catabolite-sensitive enzymes. Eur J Biochem. 1974 Nov 1;49(1):305–316. doi: 10.1111/j.1432-1033.1974.tb03835.x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wijk R., Konijn T. M. Cyclic 3', 5'-amp in Saccharomyces carlsbergensis under various conditions of catabolite repression. FEBS Lett. 1971 Mar 5;13(3):184–186. doi: 10.1016/0014-5793(71)80231-4. [DOI] [PubMed] [Google Scholar]
- Willick G. E., Morosoli R., Seligy V. L., Yaguchi M., Desrochers M. Extracellular proteins secreted by the basidiomycete Schizophyllum commune in response to carbon source. J Bacteriol. 1984 Jul;159(1):294–299. doi: 10.1128/jb.159.1.294-299.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willick G. E., Seligy V. L. Multiplicity in cellulases of Schizophyllum commune. Derivation partly from heterogeneity in transcription and glycosylation. Eur J Biochem. 1985 Aug 15;151(1):89–96. doi: 10.1111/j.1432-1033.1985.tb09072.x. [DOI] [PubMed] [Google Scholar]
- Wilson J. J., Ingledew W. M. Isolation and characterization of Schwanniomyces alluvius amylolytic enzymes. Appl Environ Microbiol. 1982 Aug;44(2):301–307. doi: 10.1128/aem.44.2.301-307.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Fukui S. Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3069–3073. doi: 10.1128/mcb.5.11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]