Abstract
Expansins are proteins that induce extension in isolated plant cell walls in vitro and have been proposed to disrupt noncovalent interactions between hemicellulose and cellulose microfibrils. Because the plant primary cell wall acts as a constraint to cell enlargement, this process may be integral to plant cell expansion, and studies of expansins have focused on their role in growth. We report the identification of an expansin (LeExp1) from tomato that exhibits high levels of mRNA abundance and is specifically expressed in ripening fruit, a developmental period when growth has ceased but when selective disassembly of cell wall components is pronounced. cDNAs closely related to LeExp1 were also identified in ripening melons and strawberries, suggesting that they are a common feature of fruit undergoing rapid softening. Furthermore, the sequence of LeExp1 and its homologs from other ripening fruit define a subclass of expansin genes. Expression of LeExp1 is regulated by ethylene, a hormone known to coordinate and induce ripening in many species. LeExp1 is differentially expressed in the ripening-impaired tomato mutants Nr, rin, and nor, and mRNA abundance appears to be influenced directly by ethylene and by a developmentally modulated transduction pathway. The identification of a ripening-regulated expansin gene in tomato and other fruit suggests that, in addition to their role in facilitating the expansion of plant cells, expansins may also contribute to cell wall disassembly in nongrowing tissues, possibly by enhancing the accessibility of noncovalently bound polymers to endogenous enzymic action.
The primary cell wall of plants has been described as a network of cellulose microfibrils embedded in a hemicellulosic polysaccharide matrix that interacts to some degree with an additional coextensive matrix of pectin and other less abundant components including structural proteins (1). In dicotyledons the predominant hemicellulose is xyloglucan and it is thought that cellulose microfibrils are coated and tethered by a framework of xyloglucan polymers (2, 3). In a turgid cell, disruption of this potentially load-bearing hemicellulose–cellulose network could provide a rate-limiting step to cell wall expansion, although an enzymic basis for wall loosening remains to be established.
Candidates for mediating hemicellulose modification as a mechanism for cell expansion include endo-1,4-β-glucanases (or “cellulases”) (4) and xyloglucan endotransglycosylases (5, 6), which have been associated with rapidly expanding tissues. Neither of these classes of enzymes, however, appears to cause extension in in vitro assays using isolated cell walls (7). Instead, a class of proteins called expansins has recently been identified that cause cell wall loosening in stress–relaxation assays but that lack detectable hydrolytic or transglycosylase activity (7–9). It has been proposed that expansins disrupt noncovalent linkages, such as hydrogen bonds, at the cellulose–hemicellulose interface thereby loosening an important constraint to turgor-driven cell expansion (9).
In addition to elongation growth, disassembly of hemicellulose also appears to be integral to wall metabolism during fruit ripening when fruit typically undergo a complex change in textural and rheological characteristics. During ripening, both the pectic and hemicellulosic polymers generally undergo substantial depolymerization and solubilization (10, 11). Much research has focused on pectin degradation, resulting from the action of the ripening-related enzyme polygalacturonase, as the key element underlying the softening process. Molecular genetic studies, however, have revealed that this process is not the primary determinant of fruit softening (12, 13) but may determine other aspects of fruit quality (14). Disassembly of the hemicellulose component of the wall during ripening is common to most fruit, although the extent varies between species (15) and most likely reflects the degradation of a mixture of polysaccharides by multiple enzymes. Xyloglucan represents the predominant hemicellulose in many fruit including tomato, where degradation is apparent during ripening in wild-type fruit but not in fruit of the rin (ripening inhibitor) tomato mutant that soften extremely slowly (16). Fruit ripening has been associated with both endo-1,4-β-glucanases (15, 17) and xyloglucan endotransglycosylases (16, 18); however, the importance of these and other as yet uncharacterized enzymes in modifying hemicellulose abundance, distribution, and interaction with other cell wall components in fruit has yet to be determined.
Expansin gene families have been identified in cucumber, rice, Arabidopsis (19), and tomato (unpublished data), suggesting that divergent isoforms may act on different components of the wall, exhibit differential developmental and environmental regulation, or tissue- and cell-specific expression. Expansins have to date been examined only in vegetative tissues. We report the cloning of an expansin from tomato that exhibits high levels of mRNA accumulation and specific expression in ripening fruit and describe the regulation of its mRNA abundance by ethylene in wild-type tomatoes and in the ripening mutants Nr (never ripe), rin (ripening inhibitor), and nor (nonripening). This suggests a novel role for expansins in developmental processes associated with changes in the cell wall architecture of nongrowing tissues.
MATERIALS AND METHODS
Plant Materials.
Fruit and vegetative tissues were harvested from field-grown (Davis, California) tomatoes (Lycopersicon esculentum cv. T5) and used as the source material in Figs. 2, 3, 4. Transgenic tomatoes expressing an 1-aminocycloprane-1-carboxylic acid (ACC) synthase antisense gene (20) were greenhouse-grown (Davis, California), and fruit used as a source of RNA for the blot in Fig. 7 (Lycopersicon esculentum cv. Ailsa Craig) were grown as described in Yen et al. (21). In all cases, plant tissues were harvested at the indicated times and stages, immediately frozen in liquid nitrogen, and stored at −80°C.
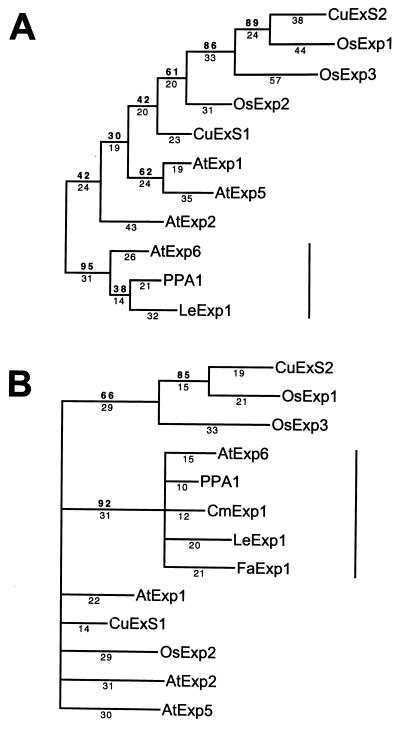
Figure 2.
(A) Phylogenetic tree of full-length deduced amino acid sequences of 11 expansins and homologs. CuExS1 and CuExS2 (Cucumis sativus); OsExp2 and OsExp3 (Oryza sativa); AtExp1, AtExp2, AtExp5, and AtExp6 (Arabidopsis thaliana) all identified in Shcherban et al. (19); PPA1 (Pisum sativum) (28); OsExp1 (Oryza sativa) EMBL accession no. Y07782Y07782. (B) Similar alignment using truncated sequences of the above genes with deduced amino acid sequences of the PCR clones CmExp1 (Cucumis melo) and FaExp1 (Fragaria ananassa) derived from melon and strawberry fruit, respectively. For each alignment, bootstrap analysis used random stepwise addition of taxa with 100 replicates and global (tree bisection and reconnection) branch swapping. Bootstrap confidence values and branch lengths are depicted above and below the lines, respectively. A vertical line represents the position of the expansin subfamily containing three ripening-related genes (LeExp1, CmExp1, and FaExp1).
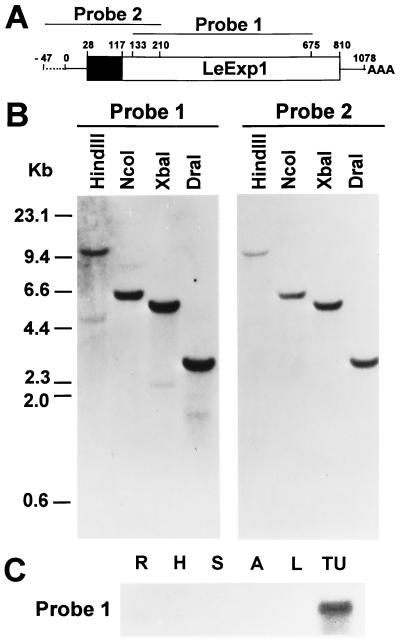
Figure 3.
(A) Diagram of the LeExp1 gene and 47 bp of 5′ flanking sequence derived from the pARC7 and pBluescript II cloning vectors. The boxed region represents the coding sequence with the solid area representing the putative signal sequence. Both 5′ and 3′ untranslated regions are depicted by solid lines and residual cloning vector sequence is depicted by a broken line. Nucleotide numbers are indicated above the gene. Two probes were designed from this sequence and used for Northern and Southern blot analyses. Probe 1 corresponded to a more conserved sequence among expansins while probe 2 corresponded to more divergent sequence. (B) Genomic DNA analysis of LeExp1. Genomic DNA (20 μg per lane) was digested with the indicated restriction enzymes and hybridized with probe 1 and washed at low stringency or with probe 2 and washed at high stringency. (C) RNA gel blot analysis of LeExp1 mRNA abundance in fruit and vegetative tissues. Total RNA (15 μg) from roots (R), hypocotyls (H), stems (S), anthers (A), young leaves (L), or turning fruit (TU) was hybridized, after gel electrophoresis, with probe 1.
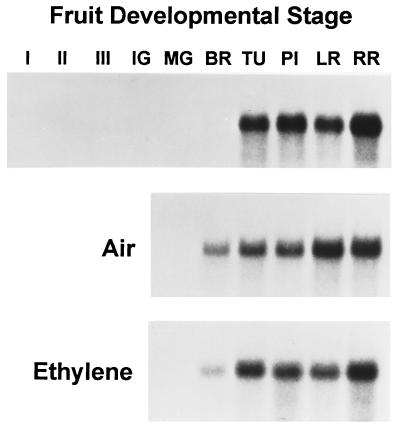
Figure 4.
Total RNA gel blot analysis of LeExp1 expression in three expanding stages of fruit (I–III), and fruit ripened on the vine (Top) or first harvested at the MG stage and allowed to ripen in the presence of air (Middle) or ethylene at 10 μl/liter (Bottom). Ripening stages included immature green (IG), MG, breaker (BR), turning (TU), pink (PI), light red (LR), and red ripe (RR). Each blot was hybridized with probe 2 and exposed to film at −80°C for approximately 9 h.
Figure 7.
RNA gel blot analysis of LeExp1 expression in fruit ripening series of wild-type fruit (Ailsa Craig) and equivalent-age rin, nor, and Nr mutant fruit. Total RNA was isolated from MG, breaker (BR), red ripe (RR), or MG fruit treated with ethylene at 20 μl/liter for 8 h (MG/E), separated by gel electrophoresis (15 μg per lane), and hybridized with probe 2. This blot was generously provided by J. Giovannoni and P. Kannan (Texas A&M University).
Ethylene Treatments.
Fruit were assigned a developmental stage based on size or color (17). Pericarp tissue was isolated from young expanding fruit (stages I, II, and III corresponding to fruit diameters of 0.5–1 cm, 2–3 cm, and 4–6 cm, respectively), vine-ripened fruit, or post-harvest-treated fruit. Mature green (MG) fruit were determined by both color and ethylene production by using a gas chromatograph fitted with a flame ionization detector. Fruit at the MG1 stage (0.02–0.1 nl of ethylene per g (fresh weight) per h) were used for subsequent continuous-flow experiments and treatments with the ethylene inhibitor 2,4-norbornadiene (NBD; Aldrich). MG1 fruit were placed in 5-liter containers and allowed to ripen in a continuous flow (20 liters/h) of humidified air or ethylene (10 μl/liter) at 25°C. Fruit were removed and tissue was frozen at the same defined stages of ripening as above. For NBD treatments, MG1 fruit were placed in sealed 20-liter chambers and held in air or NBD (2 ml/liter) with or without ethylene (10 μl/liter). Air-treated control fruit were allowed to ripen to the breaker + 4-day or red ripe + 4-day stages and on the same day tissue was collected from the NBD-treated or NBD/ethylene-treated fruit.
Flowers of the ACC synthase antisense transgenic plants were tagged at anthesis and MG fruit were harvested 37 days after pollination. Fruit were placed in 20-liter chambers and held in a continuous flow (20 liters/h) of humidified air or a defined ethylene concentration at 25°C for a period of up to 24 h.
Fruit of the wild-type Ailsa Craig cultivar, nearly isogenic and homozygous lines of the Nr and rin mutants and fruit homozygous for the nor mutation were staged and treated as in Yen et al. (21).
RNA Isolation, PCR Amplification, and cDNA Library Screening.
RNA was extracted from frozen tomato pericarp and vegetative tissues as in Rose et al. (22) and additional nucleic acid techniques used were as described in Sambrook et al. (23), unless specified otherwise.
An alignment of deduced amino acid sequences from nine expansins (19) was used to identify two conserved amino acid domains for the construction of degenerate PCR primers. The 5′ primer [G(GC)(N)CA(TC)GC(N)AC(N)TT(CT)TA(CT)GG(N)G] corresponded to amino acids 6–11 of the consensus sequence and the 3′ primer [(TC)TGCCA(AG)TT(TC)TG(N)CCCCA(AG)TT] to amino acids 182–188 (N = A, T, C, or G). cDNA synthesis from 6 μg of total RNA from turning fruit and PCR amplification with 0.5 μg of cDNA for 40 cycles (94°C for 1 min, 50°C for 1.5 min, and 72°C for 1.5 min) were as in Rose et al. (22). The resulting 542-bp cDNA fragment was gel-purified and cloned into pCR-II (Invitrogen). DNA sequence was determined with universal and specific internal primers (Genset, La Jolla, CA), using an Applied Biosystems model 377 sequencer (Perkin–Elmer) using dye terminator chemistry with AmpliTaq DNA polymerase, FS (Taq; FS; Perkin–Elmer/Applied Biosystems). The PCR fragment (probe 1) was radiolabeled by random priming using [α-32P]dATP (3,000 Ci/mmol; 1 Ci = 37 GBq; DuPont/NEN) and the Klenow fragment of DNA polymerase (United States Biochemical) and used to screen a red ripe fruit cDNA library in the pARC7 vector (24). Eight independent inserts were subcloned from the library vector into the XbaI site of the pBluescript II SK+ plasmid (Stratagene) and sequenced as for the PCR product. The longest clone was designated LeExp1. Reverse transcription-coupled PCRs similar to those described above were carried out with RNA from ripening melon and strawberry fruit.
Southern Blot Analysis.
Genomic DNA was isolated from young tomato leaves (cv. T5) as in Sambrook et al. (23), 20-μg aliquots were digested with the indicated restriction enzymes, fractionated on a 0.8% agarose gel, and transferred to Hybond-N membrane (Amersham). The blot was hybridized with probe 1 as described above. Hybridization and washing procedures were as described in Rose et al. (22) but the final three washes were at 45°C [melting temperature (Tm) − 33°C]. The blot was stripped with three washes of 0.1% SDS at 65°C and reprobed with a 257-bp radiolabeled fragment (probe 2) corresponding to nucleotides 1–210 of LeExp1 plus nucleotides 814–850 of the pARC7 and 736–745 of the pBluescript II plasmids. Hybridization was as before but the final three washes were at 60°C (Tm − 18°C).
Northern Blot Analysis.
Total RNA was isolated from all tissues as described above and 15 μg from each sample was subjected to electrophoresis on 1.2% (wt/vol) agarose/10% (vol/vol) formaldehyde denaturing gels and transferred to Hybond-N membrane. The blot used in Fig. 7 was prepared as described in Yen et al. (21). Membranes hybridized with probe 2 were washed three times at 65°C (Tm − 18°C) and the membrane hybridized with probe 1 (Fig. 3C) was washed at 45°C (Tm − 38°C). Hybridization was quantified by exposure to a PhosphorImager plate and analyzed with a Fujix BAS 1000 PhosphorImager and Fujix macbas software (Fuji).
RESULTS AND DISCUSSION
Cloning and Phylogenetic Analysis of a Tomato Expansin.
Sequence analysis of a 542-bp cDNA fragment derived by reverse transcription-coupled PCR from turning fruit RNA indicated the existence of an expansin homolog in tomato fruit (LeExp1). Subsequent screening of a red ripe tomato fruit cDNA library identified 30 positives clones, 8 of which were selected based on size, subcloned, and confirmed to have identical sequence to the original LeExp1 partial-length cDNA and to each other, but of different lengths. The longest clone (1070 bp) encoded a predicted polypeptide of 261 amino acids with an N-terminal signal sequence of 30 amino acids when the (−3, −1) rule was applied (25). An ATG codon initiated an open reading frame at position 28 and a TAA consensus stop codon was present at position 811.
A search of the GenBank database with the LeExp1 deduced amino acid sequence revealed a high degree of homology to two biochemically characterized expansins from cucumber (19) and homologs from Arabidopsis, rice, and pea. Previous analysis of these sequences identified no known functional motifs; however, it has been suggested that the N termini contain eight conserved cysteines with similar spacing to the chitin-binding domain of wheat-germ agglutinin and the C termini contain a region of conserved tryptophan residues somewhat similar to the cellulose binding domain of bacterial cellulases (19). The LeExp1 deduced amino acid sequence was aligned with five sequences from four other species, including both monocotyledons and dicotyledons (Fig. 1), and conservation of these two features was observed. A high degree of amino acid identity was apparent throughout the proposed mature polypeptides with substantial sequence divergence being evident over approximately the first 30 amino acids, corresponding to the predicted signal sequences. The high sequence identity over the entire coding sequence of LeExp1 to two cucumber expansins (CuExS1, 66%; CuExS2, 58%), a pea pollen allergen homolog isolated from pea petals (PPA1, 78%), and sequences from Arabidopsis (AtExp6, 76%) and rice (OsExp1, 56%) is of the same degree as that between the two biochemically characterized cucumber expansins (63% over the same region), suggesting that all these genes encode expansins.
Figure 1.

Multiple alignment of the LeExp1 deduced amino acid sequence with expansins and expansin homologs (see Fig. 2) using macdnasis pro 3.5 (Hitachi Software, San Bruno, CA). Amino acids conserved between any two sequences are indicated in reverse contrast and numbers above the alignment refer to numbering of the consensus sequence. Conserved tryptophan and cysteine residues are indicated by asterisks and crosses, respectively.
The above sequences and six additional homologous genes, including full-length sequences from rice and Arabidopsis, were aligned using pileup (Wisconsin Package, version 8, Genetics Computer Group, Madison, WI). A phylogram was derived (Fig. 2A) with a pollen allergen from Phleum pratense (GenBank accession no. X78813X78813) as the outgroup, using paup software (26) and bootstrap analysis. PhP1 is somewhat divergent from the other sequences (approximately 25% sequence identity); however, it retains some regions of higher homology as well as the conserved tryptophans described above, and it has been suggested that this type 1 class of allergens may function as expansins (27). LeExp1 aligned in a distinct clade with PPA1, which is expressed in pea petals (28) and AtExp6 from Arabidopsis, which has not been studied in terms of its expression patterns or biochemical properties. Other Arabidopsis sequences aligned with different branches and, as has been noted previously (19), appear more related to other sequences from both monocotyledons and dicotyledons, suggesting that divergence of these genes predated the evolutionary divergence of the angiosperms. Similar reverse transcription-coupled PCRs were carried out with RNA from ripening melon and strawberry fruit, and in each case cDNAs (CmExp1 and FaExp1 respectively) with high sequence similarity to LeExp1 were identified, suggesting that the expression of expansin genes may be a common feature of ripening fruit. To determine whether ripening-associated expansins define a subfamily of expansin genes, each of the sequences in Fig. 2A was truncated to correspond to the size of the strawberry and melon PCR fragments and aligned as described above (Fig. 2B). Alignment of this truncated domain demonstrated a phylogenetic relationship between all of the expansins similar to that observed over the entire sequence (Fig. 2A) and, furthermore, indicated that, along with PPA1 and AtExp6, the ripening-associated expansins define a subfamily of expansin genes.
Genomic Analysis and Expression of a Fruit-Ripening-Specific Expansin.
Expansin gene families of various complexities have been reported in Arabidopsis, rice, and cucumber (19, 27). Fig. 3A represents the LeExp1 cDNA clone and indicates the regions of the cDNA used to construct two probes for the determination of the potential complexity of the expansin gene family in tomato. Probe 1 (amino acid 133–675) corresponded to the central portion of the gene that is most conserved among the expansins and their homologs (Fig. 1). A Southern blot of tomato genomic DNA hybridized with probe 1 and washed at low stringency (Fig. 3B) revealed one major hybridizing band and at least two weaker bands, suggesting that LeExp1 is a member of a small multigene family. Since larger expansin gene families have been reported in other species (27), it is possible that only a subset of the total tomato expansin gene family was detected and that LeExp1 may reflect a divergent clade that does not cross-hybridize with other expansin genes (Fig. 2A). A second probe (probe 2) was designed from the more divergent 5′ portion of LeExp1 and used to probe the same Southern blot. Only the single major band that was seen with probe 1 was evident, indicating that probe 2, when used at this stringency, detected a single gene in tomato.
Both probes were used to examine expression of LeExp1 and related genes in a variety of tomato tissues at the level of mRNA abundance, at the same relative stringencies as the Southern blots. Probe 1 hybridized strongly to a 1.1-kb RNA isolated from fruit at the turning stage of ripening (Fig. 3C). After prolonged exposure of the membrane to film, a low level of hybridization (<1% of signal in turning fruit) was detected in roots, hypocotyls, stems, and young leaves. Interestingly, expression was not detected in anthers, which presumably contained a quantity of pollen, despite the homology of LeExp1 and other expansins to pollen allergens. Probe 2 detected a similar abundance of LeExp1 mRNA in turning fruit but not in other tissues, even after prolonged exposure of the blot to film (data not shown), suggesting that the expression of LeExp1 is fruit specific.
Ripening and Ethylene Regulation of LeExp1.
Fruit development from a mature ovule through final maturity encompasses a wide range of complex and highly regulated physiological processes. Early development in most fruit can be divided into three phases: fruit set, cell division, and cell expansion (29). Upon reaching full expansion ripening is initiated, typically involving changes in color, aroma, flavor, and a textural transition that contributes to softening of the tissue. The ripening process in climacteric fruit such as tomato, banana, and apple is highly regulated by the plant hormone ethylene, which is thought to coordinate the numerous metabolic pathways necessary for normal ripening. Expression of LeExp1 was examined at the level of total mRNA in fruit ripened attached to the vine or harvested prior to the onset of ripening at the MG stage and allowed to ripen off the vine in the presence of air or exogenous ethylene (10 μl/liter; Fig. 4). In vine-ripened fruit, LeExp1 was not detected in expanding or full-size nonexpanding fruit prior to the breaker stage, which marks the onset of autocatalytic ethylene production. LeExp1 mRNA was first detected at the breaker stage of fruit ripening and its abundance increased dramatically at the turning stage, remaining extremely high throughout ripening. Similar patterns of LeExp1 expression were evident in fruit ripened off the vine in the presence or absence of exogenous ethylene, suggesting that LeExp1 expression is tightly linked to ripening, since temporally the air-ripened fruit reached the same ripening stage as the ethylene-treated fruit 7–10 days later.
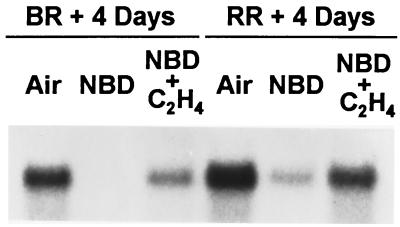
To examine the potential role of ethylene in regulating LeExp1 expression, an inhibitor of ethylene action, NBD, was used that competes with ethylene for the ethylene receptor (30). LeExp1 mRNA accumulation was abolished by NBD in fruit at breaker + 4 days and showed severely reduced levels in over-ripe fruit (Fig. 5). This effect was reversed in both stages by coincubation with ethylene, presumably due to competition for the ethylene receptor, suggesting that ethylene directly regulates LeExp1 mRNA abundance.
Figure 5.
Effect of NBD on LeExp1 expression. MG fruit were harvested and held in air, NBD at 2 ml/liter, or the same NBD concentration plus ethylene at 10 μl/liter until air-treated control fruit reached the breaker plus 4 day or red ripe plus 4 day (over ripe) stages. Fruit from all three treatments were used to isolate total RNA (15 μg per lane) for RNA gel blot analysis using probe 2.
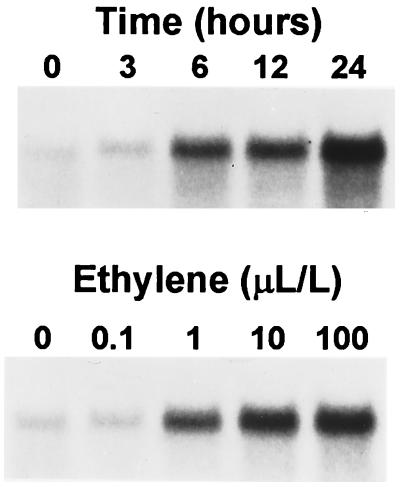
The autocatalytic nature of ethylene production during ripening complicates any determination of the threshold levels necessary to induce LeExp1 mRNA accumulation and the time frame in which induction occurs. These questions were addressed with transgenic tomatoes exhibiting a greater than 99% inhibition of ethylene production, resulting from the expression of an antisense RNA of ACC synthase (20). Transgenic fruit from these plants fail to ripen in the absence of exogenous ethylene and 6 days of continuous treatment of MG transgenic fruit with ethylene at 10 μl/liter are necessary to restore a normal phenotype (31). Expression of LeExp1 mRNA was examined in these fruit treated for 24 h with a range of ethylene concentrations or over a time course of 24 h with ethylene at 10 μl/liter (Fig. 6). Basal levels of LeExp1 mRNA were detected prior to treatment. After incubation for 24 h in a range of ethylene concentrations, the threshold of ethylene induction was seen at 0.1–1 μl/liter with little difference between treatments with ethylene at 10 μl/liter and 100 μl/liter. During a treatment of fruit with exogenous ethylene at 10 μl/liter over a 24-h time course, a large induction of LeExp1 mRNA accumulation was seen after 6 h and increased linearly throughout the 24-h treatment, suggesting that LeExp1 mRNA is relatively stable or that the transcription rate also continued to increase over 24 h. The rapid induction of LeExp1 mRNA after only 6 h of treatment with exogenous ethylene indicates that LeExp1 transcription or transcript stability is ethylene-regulated.
Figure 6.
Time course and concentration series of ethylene induction of LeExp1 in ACC synthase antisense transgenic fruit. MG fruit were harvested and treated with ethylene at 10 μl/liter for up to 24 h (Upper) or with a range of ethylene concentrations for 24 h (Lower). Total RNA (15 μg per lane) was used for RNA gel blot analysis and hybridized with probe 2.
Differential Expression of LeExp1 in the Ripening Mutants Nr, rin, and nor.
An alternative approach to dissecting the complexity and molecular basis of the ripening process has been through the study of ripening mutations, principally in the pleiotropic tomato mutants Nr (never ripe), rin (ripening inhibitor), and nor (nonripening). Nr is a dominant mutation, resulting from a single amino acid change in a homolog of the Arabidopsis ethylene receptor ETR1 (32). Fruit of the Nr mutant exhibit only partial delayed ripening and minimal softening occurs. The bases for the rin and nor mutations, both of which are recessive, are not known; however, the ripening-impaired phenotypes are more severe (33) and fruit softening is dramatically reduced (34). All three ripening-impaired mutants have been used as tools to study the processes underlying cell wall disassembly during fruit ripening, through analysis of the expression of cell wall hydrolases such as polygalacturonase (35) and endo-1,4-β-glucanases (17), and of cell wall polymer synthesis (34) and degradation (16) during ripening. By using a similar approach, the accumulation of LeExp1 mRNA was examined in MG, breaker, red ripe, and ethylene-treated MG wild-type fruit, and equivalent-age Nr, rin, and nor mutant fruit (Fig. 7). As before, high levels of LeExp1 mRNA were observed at the breaker and red ripe stages in wild-type fruit and an increase in abundance was detected in MG fruit upon ethylene treatment after prolonged exposure of the blot to film. In nor and rin fruit, basal levels of transcript (<1% and 2% of wild type, respectively) were detected and exogenous ethylene treatment of MG fruit caused no detectable induction. The low levels of LeExp1 mRNA, therefore, correlate with the previously characterized reduction in the rate of fruit softening (34) in these mutants. High levels of LeExp1 mRNA were apparent in Nr, equivalent to those in wild type; however, ethylene treatment did not induce enhanced mRNA levels. The severity of the Nr phenotype appears to depend on the genetic background, and the fruit of Nr in the Ailsa Craig background, used in these experiments, exhibit a degree of ethylene responsiveness and ripen to a greater extent than in other backgrounds (36). The possibility that high levels of LeExp1 mRNA accumulation were detected partly as a result of a leaky Nr mutation cannot be excluded.
It has been demonstrated that a variety of ripening-related genes are differentially expressed among these mutants and a model has been proposed in which the Nr gene product is necessary for regulation of most ethylene-regulated genes (21). These include genes that are regulated either primarily by ethylene or by an additional developmental component. The model further describes the rin and nor gene products as regulatory elements of a developmental pathway in which fruit acquire competence to respond to the ethylene signal, thereby playing a more indirect role in ethylene perception. The expression patterns of LeExp1 mRNA in Fig. 7 and the previous experiments suggest that LeExp1 is regulated directly by ethylene and is also influenced by a developmental pathway that appears to be modulated by the rin and nor genes. The severely reduced levels of detectable LeExp1 mRNA in the nonsoftening rin and nor mutants suggest that LeExp1 may play a role in the cell wall disassembly that occurs during fruit ripening.
CONCLUSIONS
Expansins have to date been studied only in elongating vegetative tissues and have been suggested to play a role in wall loosening, allowing turgor-driven cell expansion. However, the mechanism by which this occurs or the substrates involved in this process have not been elucidated. McQueen-Mason and Cosgrove (9) concluded that expansins bind at the interface between cellulose microfibrils and matrix polysaccharides and that this binding disrupts noncovalent interactions between these polymeric components. This suggestion is consistent with the cellulose binding domain-like motif that is conserved among expansins (Fig. 1). It has also been suggested that the interface between cellulose and xyloglucan, the major hemicellulosic polysaccharide in the wall of dicotyledons, is unlikely to represent a target for expansin action (9). The interaction between cellulose and xyloglucan is known to be complex and xyloglucan is proposed not only to coat and cross-link microfibrils but also to permeate within amorphous regions of the cellulose (4, 37). Based on this complexity of cell wall polymer interactions and the apparent structural diversity among a large family of expansin proteins, we suggest that the xyloglucan–cellulose interface should not be discounted as a potential site for expansin action.
Potential roles and sites of action of ripening-related expansins include disruption of noncovalent bonds between hemicellulose and cellulose. This could have the effect of exposing previously inaccessible structurally important polymers to the action of ripening-associated cell wall enzymes such endo-1,4-β-glucanases, xyloglucan endotransglycosylases, or glycosidases. In this model, cooperativity between expansins and these enzymes could provide a highly regulated mechanism for cell wall disassembly. It has also previously been suggested that a reduction in the degree of hydrogen bonding between xyloglucan and cellulose during fruit ripening may contribute to increased hydration of the cell wall during fruit ripening, allowing enhanced solubilization of other polymers, possibly those physically entangled in the wall (38). Alternatively, ripening-specific expansins might disrupt noncovalent linkages between other cell wall polymers, such as those between hemicelluloses and pectins. The existence of such linkages is currently hypothetical; however, small quantities of xylan–pectin complexes have been isolated from tomato fruit (39). The identification of a divergent expansin that may play a previously unrecognized role in plant development raises the possibility that additional expansin isoforms may also have a function in tissues that are not expanding but require rapid wall turnover and modification, such as abscission zones or cells undergoing nonexpansive differentiation.
The mechanism of action and in vivo function of expansins in general, and ripening-specific expansins in particular, remain uncertain. Such questions are currently being addressed, in part through the use of transgenic plants to establish whether LeExp1 plays an integral role in the cell wall disassembly during fruit ripening and whether expansin action is necessary to mediate the softening process.
Acknowledgments
We thank Prof. Jim Giovannoni and Dr. Priya Kannan (Texas A&M University, College Station) for their assistance and generous gift of the membrane shown in Fig. 7, and Prof. Athanasios Theologis (Plant Gene Expression Center, University of California at Berkeley–United States Department of Agriculture, Albany, CA) and Dr. Pam Dunsmuir (DNA Plant Technology, Oakland, CA) for supplying transgenic tomato seed and strawberry RNA, respectively. We also thank Dr. Yaron Sitrit (University of California, Davis) for providing ethylene-treated transgenic tissue, and Kristen Hadfield and Drs. Ann Powell (University of California, Davis) and Coralie Lashbrook (University of Florida, Gainsville) for helpful assistance and discussion.
ABBREVIATIONS
- NBD
2,4-norbornadiene
- MG
mature green
- ACC
1-aminocycloprane-1-carboxylic acid
Footnotes
References
- 1.Carpita N C, Gibeaut D M. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi T, Maclachlan G. Plant Physiol. 1984;75:596–604. doi: 10.1104/pp.75.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCann M C, Wells B, Roberts K. J Cell Sci. 1990;96:323–334. [Google Scholar]
- 4.Fry S C. Physiol Plant. 1989;75:532–536. [Google Scholar]
- 5.Fry S C, Smith R C, Renwick K F, Martin D J, Hodge S K, Matthews K J. Biochem J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishitani K, Tominaga R. J Biol Chem. 1992;267:21058–21064. [PubMed] [Google Scholar]
- 7.McQueen-Mason S, Durachko D M, Cosgrove D J. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McQueen-Mason S J, Fry S C, Durachko D M, Cosgrove D J. Planta. 1993;190:327–331. doi: 10.1007/BF00196961. [DOI] [PubMed] [Google Scholar]
- 9.McQueen-Mason S J, Cosgrove D J. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross K C, Wallner S J. Plant Physiol. 1979;63:117–120. doi: 10.1104/pp.63.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber D J. Hortic Rev. 1983;5:169–215. [Google Scholar]
- 12.Smith C J S, Watson C F, Ray J, Bird C R, Morris P C, Schuch W, Grierson D. Nature (London) 1988;334:724–726. [Google Scholar]
- 13.Giovannoni J J, DellaPenna D, Bennett A B, Fischer R L. Plant Cell. 1989;1:53–63. doi: 10.1105/tpc.1.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuch W, Kanczler J, Hobson G, Tucker G, Grierson D, Bright S, Bird C R. Hortscience. 1991;26:1517–1520. [Google Scholar]
- 15.Lashbrook C C, Brummell D A, Rose J K C, Bennett A B. In: Fruit Ripening Molecular Biology. Giovannoni J J, editor. Reading, U.K.: Harwood Academic; 1997. , in press. [Google Scholar]
- 16.Maclachlan G, Brady C. Plant Physiol. 1994;105:965–974. doi: 10.1104/pp.105.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Bosch C, Brummell D A, Bennett A B. Plant Physiol. 1996;111:1313–1319. doi: 10.1104/pp.111.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrowsmith D A, de Silva J. Plant Mol Biol. 1995;28:391–403. doi: 10.1007/BF00020389. [DOI] [PubMed] [Google Scholar]
- 19.Shcherban T Y, Shi J, Durachko D M, Guiltinan M J, Shieh M, Cosgrove D J. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oeller P W, Wong L M, Taylor L P, Pike D A, Theologis A. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- 21.Yen H, Lee S, Tanksley S D, Lanahan M B, Klee H J, Giovannoni J J. Plant Physiol. 1995;107:1343–1353. doi: 10.1104/pp.107.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose J K C, Brummell D A, Bennett A B. Plant Physiol. 1996;110:493–499. doi: 10.1104/pp.110.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.DellaPenna D, Kates D S, Bennett A B. Proc Natl Acad Sci USA. 1986;83:6420–6424. doi: 10.1073/pnas.83.17.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford D. Illinois Natural History Survey. IL: Champaign; 1993. [Google Scholar]
- 27.Cosgrove D J. BioEssays. 1996;18:533–540. doi: 10.1002/bies.950180704. [DOI] [PubMed] [Google Scholar]
- 28.Michael A J. Plant Mol Biol. 1996;30:219–224. doi: 10.1007/BF00017818. [DOI] [PubMed] [Google Scholar]
- 29.Gillaspy G H, Ben-David H, Gruissem W. Plant Cell. 1993;5:1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sisler E C, Yang S F. Phytochemistry. 1984;23:2765–2768. [Google Scholar]
- 31.Theologis A. Cell. 1992;70:181–184. doi: 10.1016/0092-8674(92)90093-r. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson J O, Lanahan M B, Yen H, Giovannoni J J, Klee H J. Science. 1995;270:1807–1808. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 33.Tigchelaar E C, McGlasson W B, Buescher R W. Hortscience. 1978;13:508–513. [Google Scholar]
- 34.Mitcham E J, Gross K C, Ng T J. Phytochemistry. 1991;30:1777–1780. [Google Scholar]
- 35.DellaPenna D, Kates D S, Bennett A B. Plant Physiol. 1987;85:502–507. doi: 10.1104/pp.85.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanahan M B, Yen H, Giovannoni J J, Klee H J. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi T. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- 38.Redgwell R J, Melton L D, Brasch D J. Carbohydr Res. 1991;209:191–202. [Google Scholar]
- 39.Seymour G B, Colquhoun I J, Dupont M S, Parsley K R, Selvendran R R. Phytochemistry. 1990;29:725–731. [Google Scholar]