Abstract
Ocs elements are a group of promoter sequences required for the expression of both pathogen genes in infected plants and plant defense genes. Genes for ocs element binding factors (OBFs), belonging to a specific class of basic-region leucine zipper (bZIP) transcription factors, have been isolated in a number of plants. Using protein–protein interaction screening with OBF4 we have isolated AtEBP, an Arabidopsis protein that contains a novel DNA-binding domain, the AP2/EREBP domain. One class of proteins that contain this domain are the tobacco ethylene-responsive element binding proteins (EREBPs). The EREBPs bind the GCC box that confers ethylene responsiveness to a number of pathogenesis related (PR) gene promoters. AtEBP expression is inducible by exogenous ethylene in wild-type plants and AtEBP transcripts are increased in the ctr1-1 mutant, where ethylene-regulated pathways are constitutively active. Electrophoretic mobility-shift assay and DNase I footprint analysis revealed that AtEBP can specifically bind to the GCC box. Interestingly, the highest level of AtEBP expression was detected in callus tissue, where ocs elements are very active. Synergistic effects of the GCC box with ocs elements or the related G-box sequence have been previously observed, for example, in the ethylene-induced expression of a PR gene promoter. Our results suggest that cross-coupling between EREBP and bZIP transcription factors occurs and may therefore be important in regulating gene expression during the plant defense response.
Keywords: ethylene, gene regulation, plant defense, pathogenesis related genes
Pathogenesis-related (PR) proteins are a diverse group of proteins that have been linked to the plant defense response (reviewed in refs. 1 and 2). PR proteins were first discovered in tobacco where they accumulate in leaves following infection of susceptible genotypes with tobacco mosaic virus (3, 4). Subsequently, PR proteins have been found in a number of plant species. While some PR proteins such as chitinases and β-1,3-glucanases have known catalytic functions, the functions of other PR proteins remain to be determined. A number of studies have supported a role for PR proteins in the plant defense response (reviewed in refs. 5 and 6). Thus, PR proteins have been shown to inhibit bacterial and fungal growth in vitro, while overexpression of PR proteins in transgenic plants resulted in increased resistance to some fungal pathogens. Interestingly, evidence for synergistic activity has been found for chitinases and β-1,3-glucanases from both in vitro (7) and in vivo studies (8).
In addition to pathogen attack, the expression of specific PR genes is induced by other stimuli such as UV, salicylic acid, and ethylene (see ref. 9 and references within). Both salicylic acid, a plant defense signal, and ethylene accumulate in plants during pathogen infection. Analysis of PR gene promoters has led to the identification of an 11-bp ethylene-responsive element, TAAGAGCCGCC (10–13), which has been referred to as the GCC box (12). Four ethylene-responsive element binding proteins (EREBPs) have been isolated in tobacco that contain a novel DNA-binding domain and specifically bind to the GCC box (12). The EREBP RNA levels are up-regulated by ethylene, suggesting that EREBP expression may also be induced during the defense response. Further evidence linking EREBPs with the defense response has come from an analysis of a tomato resistance (R) gene, Pto (14). The R genes confer gene-for-gene resistance by activating the plant defense response following specific recognition of a plant pathogen (15). Pto-interacting proteins that resemble the tobacco EREBPs have been isolated in tomato, suggesting a possible mechanism for the regulation of PR gene expression by EREBPs through direct interaction with R gene products (reviewed in ref. 15).
A second class of transcription factors that may play a role in the plant defense response are ocs element binding factor (OBF) proteins (16, 17), which bind to a family of related, 20-bp DNA promoter sequences called ocs elements (18). The OBF proteins belong to a specific class of highly conserved, plant basic-region leucine zipper (bZIP) transcription factors that include the tobacco and Arabidopsis TGA proteins (19). Reverse genetic experiments (20) and expression studies (21, 22) have provided strong evidence linking this family of bZIP proteins with ocs element activity. Both bacterial and viral pathogens utilize ocs elements to express genes in plants (18). Ocs elements also regulate transcription of plant glutathione S-transferase (GST) genes (23, 24) and studies have linked GST genes with the plant defense response (25–29). To further analyze the ocs element, we have attempted to isolate OBF interacting proteins by screening a cDNA library with the Arabidopsis OBF4 protein. In this report, we describe the isolation and characterization of AtEBP, an Arabidopsis thaliana ethylene-responsive element binding protein.
METHODS
Expression and Labeling of Recombinant Proteins.
OBF4 was prepared and labeled as described (30). AtEBP was overexpressed in Escherichia coli as a GST fusion protein using the pGEX–2TK plasmid vector (Pharmacia). A fragment containing the entire coding region of AtEBP was produced using the PCR and the oligonucleotide sequences listed below and was inserted into the BamHI site of pGEX–2TK. The primer sequences for AtEBP are as follows: 5′-CGGGATCCATGTGTGGCGGTGCTATTATTTCCG-3′ and 5′-GGGGATCCTCATACGACGCAATGACATC-3′.
The pGEX–2TK constructs with or without AtEBP were transformed into E. coli BL21(DE3) competent cells. Five milliliters of an overnight culture was added to 500 ml of Luria-Bertani medium and grown for 3 hr at 37°C. The expression of the GST fusion proteins was then induced for 2 hr with the addition of 0.5 mM isopropyl β-d-thiogalactoside. The purification of the GST-fusion proteins using glutathione beads, and the digestion with thrombin (Sigma), was as described (31).
Library Screening and DNA Sequence Analysis.
Labeled OBF4 was used to screen 240,000 plaques from an Arabidopsis callus cDNA expression library (17). The protein screening technique was as described (30). Screening of the callus library to isolate larger AtEBP clones was performed using standard techniques. Single-stranded DNA sequence reactions were performed using the Sequenase Version 2.0 DNA sequencing kit (Amersham) and the plasmid vector pBluescript SK+ (Stratagene).
Protein–Protein Binding Studies.
One hundred nanograms of labeled OBF4 protein was incubated in 20 μl PBS buffer for 30 min at room temperature with either glutathione-agarose beads alone or beads with bound GST or GST–AtEBP. After washing for three times with 1 ml PBS, the bound protein was recovered by elution in 5 mM glutathione in 50 mM Tris (pH 7.5) and analyzed by SDS/PAGE followed by autoradiography.
DNA and RNA Gel Blot Analysis.
Genomic DNA was extracted from Arabidopsis thaliana (ecotype Columbia) based on the method of Dellaporta et al. (32). Genomic DNA (10 μg) was digested with KpnI, HindIII, EcoRV, EcoRI, or BamHI. DNA gel blot analysis was performed using standard techniques and a random-primed AtEBP cDNA probe. Total RNA (5 or 15 μg) from Arabidopsis seedlings or callus tissue was fractionated on a 1.5% agarose-formaldehyde gel with RNA molecular mass markers (Bethesda Research Laboratories). RNA gel blot analysis was as described (17), except that a random primed AtEBP cDNA probe was used. For the different treatments 8-day-old seedlings were incubated for 2 hr in Murashige and Skoog (MS) medium with or without 5 mM hydrogen peroxide, for 12 hr in MS medium with different concentrations of cadmium or copper or for 12 hr in phosphate buffer (pH 7.0) containing 1 mM ethephon (2-chloroethyl-phosphonic acid) or 1 mM kinetin.
Electrophoretic Mobility-Shift Assay and DNase I Footprinting Assays.
The electrophoretic mobility-shift assay and DNase I footprinting experiments were performed with AtEBP overexpressed and purified from E. coli. For the GCC box DNA probes the oligonucleotides shown in Fig. 6 were synthesized with flanking BamHI and BglII sites and cloned into the BamHI site of the pBluescript polylinker as described (33). XbaI/BamHI fragments containing the oligonucleotides were gel purified, end-labeled, and used in gel retardation experiments as described (30). For the footprint analysis the −209/−173 promoter fragment of the PRB-1b promoter was cloned into the BamHI site of pBluescript SK+. This plasmid served as template for the PCRs using the T7 and the end-labeled T3 primer and vice versa, to give a 200-bp fragment that was gel-purified. The binding reaction, DNase I digest, and gel analysis were as described (34). The G+A sequencing ladder was generated by performing Maxam–Gilbert chemical reactions using the same 200-bp fragment.
Figure 6.
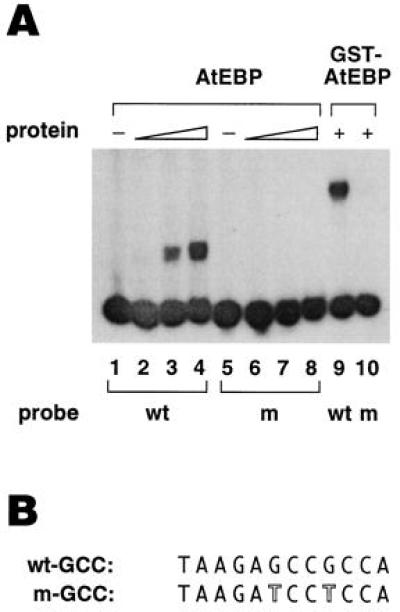
Analysis of the DNA-binding properties of AtEBP. (A) Analysis of AtEBP binding to the GCC wild-type (wt) sequence from the tobacco PRB-1b promoter (−201 to −191) and a GCC mutant (m) sequence. Lanes 1 and 5 contained only the free probes. Lanes 2 and 6 contained 5 ng, lanes 3 and 7 contained 25 ng, and lanes 4 and 8 contained 50 ng of AtEBP. Lanes 9 and 10 contained 100 ng of GST–AtEBP. (B) The oligonucleotide sequences for the GCC wild-type and mutant probes. The mutated residues in the GCC mutant probe are outlined. The 5′ end of each probe contained a BamHI site.
RESULTS
Isolation of AtEBP.
To isolate OBF interacting proteins an Arabidopsis callus cDNA expression library was screened with labeled OBF4 as described in Zhang et al. (30). A callus library was used because the ocs element is very active in callus tissue (35). Here, we focus on one of the isolated clones, called AtEBP. The initial screening led to the isolation of a 174-bp long clone (Fig. 1; nucleotides 39–212) containing an uninterrupted ORF. To obtain larger cDNA clones, the same library was screened using the 174-bp fragment as a probe. The largest clone was used for DNA sequence analysis. As shown in Fig. 1, this AtEBP cDNA is 923 bp long and contains an ORF of 248 amino acids. The AtEBP protein is hydrophilic and the carboxyl-terminal half has an overall negative charge of −19. There is also a stretch of basic amino acids, KRRKRK, which resembles a simian virus 40 type nuclear localization signal (36), and a proline-rich region from amino acids 140 to 159. The cDNA clone does not contain a poly(A) tail and based on the size of the AtEBP transcript obtained from RNA gel blot analysis, it is likely to be missing approximately 200 bp of the 5′ and/or 3′ untranslated sequences.
Figure 1.

Sequence of the AtEBP cDNA clone and the predicted amino acid sequence. The predicted amino acid sequence for the longest ORF is shown directly below the nucleotide sequence. A putative nuclear localization signal is underlined. The AtEBP GenBank database accession number is Y09942Y09942.
The Interaction Between OBF4 and AtEBP Can Be Detected Using an in Vitro Assay.
We analyzed the interaction between AtEBP and OBF4 using a GST pull-down assay. The AtEBP protein was expressed as a GST fusion in E. coli. The OBF4 protein was expressed as a GST fusion in the baculovirus expression system, and the GST peptide was subsequently removed by thrombin. The labeled OBF4 (OBF4*) and glutathione-agarose beads were incubated together for 30 min at room temperature either alone or in the presence of free GST protein or the GST–AtEBP fusion protein. After extensive washing of the beads, the bound protein was eluted with glutathione and analyzed by SDS/PAGE. The results shown in Fig. 2 demonstrate that OBF4 interacts with AtEBP in vitro. OBF4 alone (Fig. 2, lane 1) has a background level of binding to the glutathione-agarose beads. In the presence of GST (Fig. 2, lane 2), there was a slight increase in OBF4 binding. However, in the presence of GST–AtEBP (Fig. 2, lane 3), the amount of OBF4 associated with the glutathione-agarose beads was significantly increased.
Figure 2.
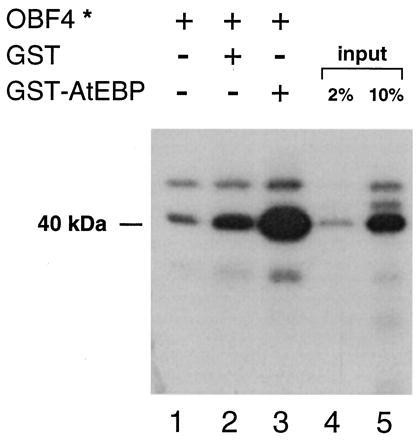
AtEBP and OBF4 interact in an in vitro assay. One hundred nanograms of labeled OBF4 protein (OBF4*) was incubated with equal amounts of either glutathione-agarose beads or beads where the GST or GST–AtEBP proteins were already bound. The amount of OBF4* protein (40 kDa) that was retained after washing the glutathione-agarose beads was purified, analyzed by SDS/PAGE, and then detected by autoradiography. Lanes 1 and 2 reflect nonspecific binding of OBF4*; lane 3 reflects the specific binding of OBF4* to GST–AtEBP bound to the glutathione-agarose beads. Aliquots of the OBF4* input are shown in lane 4 (2%) and lane 5 (10%).
AtEBP Shares Homology with Plant DNA-Binding Proteins.
Database searches were performed with both the AtEBP cDNA and predicted protein sequences. The strongest homology was found to an unpublished clone called ATCADINP (Z37504), listed as a cadmium-induced clone. AtEBP has 98% identity with two regions of the ATCADINP sequence; from nucleotides 1 to 627 and from nucleotides 834 to 962. ATCADINP has an additional 207-bp sequence (628–833) that is not found in AtEBP. However, the additional sequence in ATCADINP appears to be a duplication of sequences from nucleotides 414 to 623. The AtEBP sequence also shares very high homology with several clones from the Arabidopsis Expressed Sequence Tag (EST) database. A 57-amino acid stretch of AtEBP shares homology with other plant proteins, some of which are shown in Fig. 3. The strongest homology (71–75% identity) was found to the DNA-binding domain of the tobacco EREBPs, which are responsive to ethylene and bind to the GCC sequence motif found in the promoters of a number of PR genes. A related domain is also found in another group of plant proteins, although the degree of identity of these proteins to AtEBP is lower. This group includes proteins involved in different aspects of plant development such as APETALA2.
Figure 3.
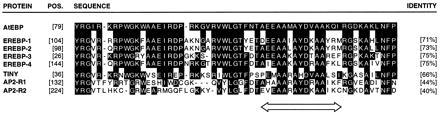
AtEBP has homology to plant DNA-binding proteins. Comparison of a region of the predicted amino acid sequence of the Arabidopsis AtEBP protein to the DNA-binding domain of the tobacco EREBPs (D38123, D38126, D38124, D38125), and similar sequences present in the Arabidopsis TINY (X94698) and APETALA2 (U12546) proteins using the Genetics Computer Group gap program. Identical amino acids are marked by black boxes. A region with a high potential for forming an amphipathic helix is indicated by a double arrow.
Analysis of the AtEBP Gene Copy Number and Expression Patterns.
We analyzed the size of the Arabidopsis AtEBP gene family by Southern blot analysis. As shown in Fig. 4, there is only a single band that hybridizes strongly with the AtEBP cDNA clone, indicating that AtEBP is a single copy gene. A number of weakly hybridizing bands were also observed. Upon longer exposure, additional hybridizing bands were observed (data not shown), suggesting that AtEBP is a member of a family of related genes in Arabidopsis.
Figure 4.
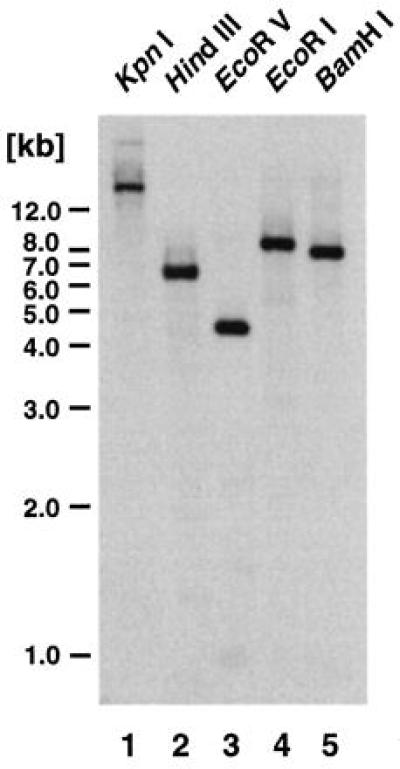
Analysis of the AtEBP gene family in Arabidopsis. Southern bolt analysis of Arabidopsis genomic DNA using the AtEBP cDNA clone as a probe. Each lane contained 10 μg of genomic DNA that was digested with the indicated enzyme. At left, a 1-kb DNA ladder (Bethesda Research Laboratories) was used for the molecular length markers.
To determine the size of the AtEBP transcript we performed Northern blot analysis with total RNA from Arabidopsis seedlings and callus tissue. We could detect a low level of AtEBP mRNA with a size of ≈1,100 bp in seedlings (Fig. 5A; lane 1). Interestingly, very high expression levels of AtEBP mRNA were detected in callus tissue (Fig. 5A; lane 2). We also tested the effect of heavy metal and other stress factors on AtEBP expression in young seedlings. Fig. 5B shows that treatment with Cd (Fig. 5B, lanes 2 and 3), Cu (lane 4) or hydrogen peroxide (lane 6) did not increase the abundance of the AtEBP mRNA compared with control seedlings.
Figure 5.
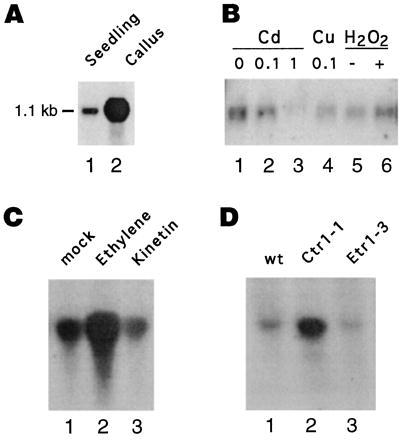
Analysis of AtEBP RNA expression patterns RNA gel blot analysis with the AtEBP cDNA clone as a probe was performed using 5 μg (A and B) or 15 μg (C and D) of total RNA from 8-day-old Arabidopsis seedlings or callus tissue. Equal amounts of RNA were loaded as determined by ethidium bromide staining. (A) Seedlings compared with callus tissue. (B) Seedlings following treatment with the indicated amounts (mM) of cadmium (Cd), copper (Cu) or hydrogen peroxide (H2O2). (C) Seedlings following treatment with phosphate buffer pH 7.0 (lane 1), or phosphate buffer pH 7.0 containing 1 mM ethephon (lane 2) or 1 mM kinetin (lane 3). (D) Wild-type (lane 1) compared with the mutant seedlings: ctr1-1 (lane 2) and etr1-3 (lane3).
Because AtEBP contains an EREBP domain, we analyzed the inducibility of AtEBP expression by ethylene in young seedlings. As shown in Fig. 5C, the addition of 1 mM ethephon increased AtEBP expression (lane 2) when compared with the treatment without any hormone (lane 1), whereas kinetin did not affect AtEBP expression (lane 3). We also tested the hormone auxin and the defense signal, salicylic acid, but neither one changed the expression levels of AtEBP (data not shown). Because exogenous ethylene induced AtEBP expression, we analyzed AtEBP expression in some of the Arabidopsis ethylene mutants. We found that the ctr1-1 mutant, where ethylene-regulated pathways are constitutively active, had increased levels of AtEBP expression (Fig. 5D, lane 2), whereas the etr1-3 mutant, which is defective in ethylene perception, had similar levels of AtEBP mRNA (lane 3) compared with wild-type plants (lane 1).
AtEBP Binds Specifically to the GCC Motif in the PRB-1b Promoter.
To analyze the DNA-binding properties of AtEBP we overexpressed the protein as a GST fusion in E. coli. After removal of the GST peptide we tested whether the AtEBP protein was able to bind specifically to the GCC box. As shown in Fig. 6, the AtEBP protein bound to a 29-bp oligonucleotide probe containing the GCC box. In some experiments the binding of AtEBP resulted in a single retarded band, although in other cases, a doublet consisting of two, closely spaced bands was observed. Whether a single or double band was observed, the intensity of the bands directly correlated with the amount of AtEBP present (Fig. 6A, lanes 2–4). We also analyzed AtEBP binding to an oligonucleotide probe containing a mutant GCC box. The mutant GCC box contained two point mutations which eliminated/reduced GCC box activity in vivo (12). The binding of AtEBP to the mutant GCC box (see Fig. 6B) was abolished (Fig. 6A, lanes 5–7). However, with high levels of AtEBP, weak binding to the mutant probe could be observed (data not shown). Similarly, the GST–AtEBP fusion protein was able to bind to the GCC box but not to the mutant version (Fig. 6A, lanes 9 and 10).
To further characterize the interaction of AtEBP with the GCC box we performed DNase I footprint analysis on both strands of a 36-bp fragment of the PRB-1b promoter (−209 to −173), which contains a GCC box. The labeled promoter fragments were digested with DNase I in the presence or absence of the AtEBP–GST fusion protein and analyzed on a 8% sequencing gel. As shown in Fig. 7, regions encompassing the GCC box were protected on both strands with AtEBP. On the bottom strand, the protected region was from −207 to −194 (Fig. 7, lanes 3 and 4), while on the top strand, the protected region was from −203 to −192 (Fig. 7, lanes 8 and 9). In both cases, the protected region was flanked by DNase I hypersensitive sites.
Figure 7.
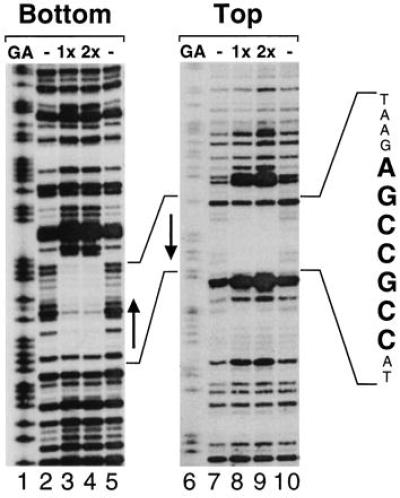
DNase I footprint analysis of AtEBP binding to the GCC box in the PRB-1b promoter. A PRB-1b promoter fragment corresponding to nucleotides −205 to −155 was labeled on both strands (bottom, top) and incubated with no protein (lanes 2, 5, 7, and 10), 50 ng AtEBP (1x; lanes 3 and 8), or 100 ng AtEBP (2x; lanes 4 and 9). The reactions were analyzed on a 8% sequencing gel together with G+A sequencing ladders generated from the same PRB-1b promoter fragments. The GCC box on the PRB-1b promoter is indicated by arrows.
DISCUSSION
By protein–protein interaction screening with OBF4 we have isolated an Arabidopsis GCC box-binding protein called AtEBP. The GCC box, also referred to as the AGC box (10), GCC element (11), or AGCCGCC sequence (13), is an ethylene-responsive element found in the promoters of a large number of PR genes whose expression is up-regulated following pathogen attack. However, the GCC box has not been found in the promoters of ethylene-regulated genes involved in other ethylene responses, such as fruit ripening (37). AtEBP shares strong homology with the DNA-binding domains of a group of tobacco GCC box-binding proteins called EREBPs (12). However, there is no homology between AtEBP and the tobacco EREBPs outside the EREBP DNA-binding domain. While further studies will be required to determine the range of DNA-binding sites for AtEBP and related EREBPs, the 11-bp GCC box is very conserved in PR gene promoters, suggesting it may be an optimal site, at least for some of these proteins.
Southern blot analysis revealed that there are a number of AtEBP-related genes in Arabidopsis. Two Arabidopsis genes, APETALA2 (38) and TINY (39), that contain the EREBP or related AP2 domain (see Fig. 2), have been isolated and characterized. APETALA2 and TINY have been proposed to be transcription factors based on the presence of the AP2/EREBP DNA-binding domain and regions that resemble transcription activation domains. The APETALA2 gene, which contains two copies of the AP2 domain, plays a regulatory role in floral organ specification and seed development (see ref. 38 and references within). The tiny mutant results from insertion of a transposable element that leads to increased expression of TINY (39). The tiny mutant displays pleiotropic phenotypes that result from changes in cell shape and cell expansion leading to a reduction in plant size (39). Some of the tiny phenotypes are similar to those observed following treatment with exogenous ethylene or with the ctr1 mutant, where ethylene signaling pathways are constitutively active. The putative DNA-binding domain in TINY is more closely related to AtEBP and the tobacco EREBPs than to either of the AP2 domains in APETALA2. These results suggest a possible link of TINY with specific ethylene responses.
The AP2/EREBP domains are novel DNA-binding domains that so far have only been found in higher plants. A region of the AP2/EREBP domain (marked in Fig. 3) has been proposed to form an amphipathic helix (38, 40). Such a helix could play a role in protein–protein interactions to facilitate DNA binding. From our binding studies we cannot determine if AtEBP binds to the GCC box as a monomer or some form of multimer. While we have occasionally resolved two, closely spaced bands, we do not feel that this reflects different oligomeric forms of AtEBP because the ratio of each band was not altered with changes in AtEBP concentrations. The double band may result from overexpression of AtEBP in E. coli or variability in our binding experiments. AtEBP also contains proline rich and acidic regions that may function as activation domains, and a putative nuclear localization sequence.
We observed higher AtEBP RNA levels in the ctr1-1 mutant and in wild-type plants following ethylene treatment. The tobacco EREBP RNA levels are up-regulated by ethylene to different degrees (12). It is not known if the expression of AtEBP or any of the tobacco EREBP genes are regulated through GCC box sequences. Interestingly, there were very high levels of AtEBP RNA in callus tissue as compared with seedlings. This observation is potentially relevant to the AtEBP/OBF4 interaction, since ocs elements are very active in Arabidopsis callus (35). Whether AtEBP expression in callus is due to elevated ethylene levels or other factors is not clear. There are conflicting reports on ethylene production in callus cultures and the amount of ethylene produced appears to be dependent on the culturing conditions (41). Chitinases have been shown to be inducible by ethylene in yam callus (42), suggesting that ethylene levels are not saturating in this tissue type. This is supported by the finding that only one of the four tobacco EREBPs is expressed in suspension culture, although all four are ethylene-inducible in leaves (12). Interestingly, an unpublished potato clone (GenBank accession no. U77655U77655) isolated from suspension cells, contains an EREBP domain and shares limited homology with AtEBP outside the EREBP domain. It will be interesting to see what developmental or tissue-specific stages are reflected in the callus that cause the high expression of AtEBP.
Cross-coupling of transcription factors is thought to play an important role in mediating responses to various signaling events; in animals for example, cross-coupling is important in modulating the transcriptional activity of members of the steroid receptor family and Jun/Fos proteins (43). Plant promoters containing both a GCC box and an ocs element have not been identified. However, a synergistic effect of GCC box containing sequences and an ocs element has been observed with an artificial promoter (10). In these experiments, two copies of a 61-bp fragment of the tobacco β-1,3-glucanase promoter, each containing two GCC boxes, were shown to synergistically interact with the ocs element from the cauliflower mosaic virus (CaMV) 35S promoter in tobacco protoplasts. A potential target for cross-coupling between EREBP and bZIP proteins could be the tobacco PRB-1b gene, whose promoter contains a G-box located 4 bp downstream of the GCC box. The G box resembles an ocs element half site and is the binding site for the G-box-binding factor family of plant bZIP proteins (reviewed in ref. 44). Both the GCC box and G box participate in ethylene-induced expression of PRB-1b (11). A similar organization of GCC-box and G-box sequences is also found in the promoters of several other PR genes including two tobacco β-glucanases and a tomato chitinase.
It will be important to analyze if AtEBP and/or related EREBP proteins can interact with the other Arabidopsis OBF/TGA proteins or with other Arabidopsis bZIP proteins such as the GBFs (45). The initial AtEBP cDNA clone isolated by the protein–protein interaction screening only contained amino acids 9–65. Therefore, this region is sufficient for the interaction between AtEBP and OBF4, at least under the conditions used in the protein–protein interaction screening. However, it is possible that other regions of AtEBP also participate in this interaction and further studies will be required to determine the precise domains responsible for the interaction between AtEBP and OBF4 as well as if this interaction results in cooperative binding. Such studies may help determine to what extent cross-coupling between EREBPs and bZIP proteins plays a role in the plant defense response and/or specific stages of plant development.
Acknowledgments
We thank Drs. B. Zhang and R. Foley for help with the protein–protein interaction screening and the RNA analysis, and Drs. R. Foley and Yiwen Fang for critical comments on the manuscript. M.B. was supported in part by a research fellowship from the Deutsche Forschungsgemeinschaft. This work was supported by a grant from the National Institutes of Health (R29-GM47893).
ABBREVIATIONS
- EREBP
ethylene-responsive element binding protein
- GST
glutathione S-transferase
- PR
pathogenesis-related
- OBF
ocs element binding factor
- bZIP
basic-region leucine zipper
- AtEBP
Arabidopsis thaliana ethylene-responsive element binding protein.
Footnotes
References
- 1.Bol J F, Linthorst H J M, Cornelissen B J C. Annu Rev Phytopathol. 1990;28:113–138. [Google Scholar]
- 2.Bowles D J. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 3.Gianinazzi S, Martin C, Vallée J C. C R Acad Sci Ser III. 1970;270:2382–2386. [PubMed] [Google Scholar]
- 4.Van Loon L C, Van Kammen A. Virology. 1970;40:199–211. doi: 10.1016/0042-6822(70)90395-8. [DOI] [PubMed] [Google Scholar]
- 5.Hammond-Kosack K E, Jones J D. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mauch F, Mauch-Mani B, Boller T. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Q, Maher E A, Masoud S, Dixon R A, Lamb C J. Bio/Technology. 1994;12:807–812. [Google Scholar]
- 9.Green R, Fluhr R. Plant Cell. 1995;7:203–212. doi: 10.1105/tpc.7.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart C M, Nagy F, Meins F., Jr Plant Mol Biol. 1993;21:121–131. doi: 10.1007/BF00039623. [DOI] [PubMed] [Google Scholar]
- 11.Sessa G, Meller Y, Fluhr R. Plant Mol Biol. 1995;28:145–153. doi: 10.1007/BF00042046. [DOI] [PubMed] [Google Scholar]
- 12.Ohme-Takagi M, Shinshi H. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato F, Katajima S, Koyama T, Yamada Y. Plant Cell Physiol. 1996;37:249–255. doi: 10.1093/oxfordjournals.pcp.a028939. [DOI] [PubMed] [Google Scholar]
- 14.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 15.Bent A F. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley R C, Grossman C, Ellis J G, Llewellyn D J, Dennis E S, Peacock W J, Singh K B. Plant J. 1993;3:669–679. [PubMed] [Google Scholar]
- 17.Zhang B, Foley R C, Singh K B. Plant J. 1993;4:711–716. doi: 10.1046/j.1365-313x.1993.04040711.x. [DOI] [PubMed] [Google Scholar]
- 18.Katagiri F, Chua N H. Trends Genet. 1992;8:22–27. doi: 10.1016/0168-9525(92)90020-5. [DOI] [PubMed] [Google Scholar]
- 19.Rieping M, Fritz M, Prat S, Gatz C. Plant Cell. 1994;6:1087–1098. doi: 10.1105/tpc.6.8.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katagiri F, Yamazaki K, Horikoshi M, Roeder R G, Chua N-H. Genes Dev. 1990;4:1899–1909. doi: 10.1101/gad.4.11.1899. [DOI] [PubMed] [Google Scholar]
- 21.Neuhaus G, Neuhaus-Uri G, Katagiri F, Seipel K, Chua N-H. Plant Cell. 1994;6:827–834. doi: 10.1105/tpc.6.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouchez D, Tokuhisa J G, Llewellyn D J, Dennis E S, Ellis J G. EMBO J. 1989;8:4197–4204. doi: 10.1002/j.1460-2075.1989.tb08605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis J G, Tokuhisa J G, Llewellyn D J, Bouchez D, Singh K, Dennis E S, Peacock W J. Plant J. 1993;4:433–443. doi: 10.1046/j.1365-313x.1993.04030433.x. [DOI] [PubMed] [Google Scholar]
- 24.Ulmasov T, Hagen G, Guilfoyle T. Plant Mol Biol. 1994;26:1055–1064. doi: 10.1007/BF00040688. [DOI] [PubMed] [Google Scholar]
- 25.Dudler R, Hertig C, Rebmann G, Bull J, Mauch F. Mol Plant–Microbe Interact. 1991;4:14–18. doi: 10.1094/mpmi-4-014. [DOI] [PubMed] [Google Scholar]
- 26.Greenberg J T, Guo A, Klessig G F, Ausubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 27.Hahn C, Strittmatter G. Eur J Biochem. 1994;226:619–626. doi: 10.1111/j.1432-1033.1994.tb20088.x. [DOI] [PubMed] [Google Scholar]
- 28.Leonards-Schippers C, Gieffers W, Schäfer-Pregl R, Ritter E, Knapp S J, Salamini F, Gebhardt C. Genetics. 1994;137:67–77. doi: 10.1093/genetics/137.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Chen W, Foley R C, Büttner M, Singh K B. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. pp. 16.7.1–16.7.8. [Google Scholar]
- 32.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 33.Singh K, Dennis E S, Ellis J G, Llewellyn D J, Tokuhisa J G, Wahleithner J A, Peacock W J. Plant Cell. 1990;2:891–903. doi: 10.1105/tpc.2.9.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen W, Chao G, Singh K B. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Singh K B. Proc Natl Acad Sci USA. 1994;91:2507–2511. doi: 10.1073/pnas.91.7.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalderon D, Richardson W D, Markham A F, Smith A E. Nature (London) 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 37.Cordes S, Deikman J, Margossian J, Fisher R L. Plant Cell. 1989;1:1025–1034. doi: 10.1105/tpc.1.10.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jofuku K D, den Boer B G W, Van Montagu M, Okamuro J K. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson K, Long D, Swinburne J, Coupland G. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weigel D. Plant Cell. 1995;7:388–389. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huxter T J, Reid D M, Thorpe T A. Physiol Plant. 1979;46:374–380. [Google Scholar]
- 42.Koga D, Hirata T, Sueshige N, Tanaka S, Ide A. Biosci Biotechnol Biochem. 1992;56:280–285. [Google Scholar]
- 43.Schule R, Evans R M. Trends Genet. 1991;7:377–381. doi: 10.1016/0168-9525(91)90259-s. [DOI] [PubMed] [Google Scholar]
- 44.Menkens A, Schindler U, Cashmore T. Trends Biochem. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- 45.Schindler U, Menkens A E, Beckmann H, Ecker J R, Cashmore A R. EMBO J. 1992;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


