Abstract
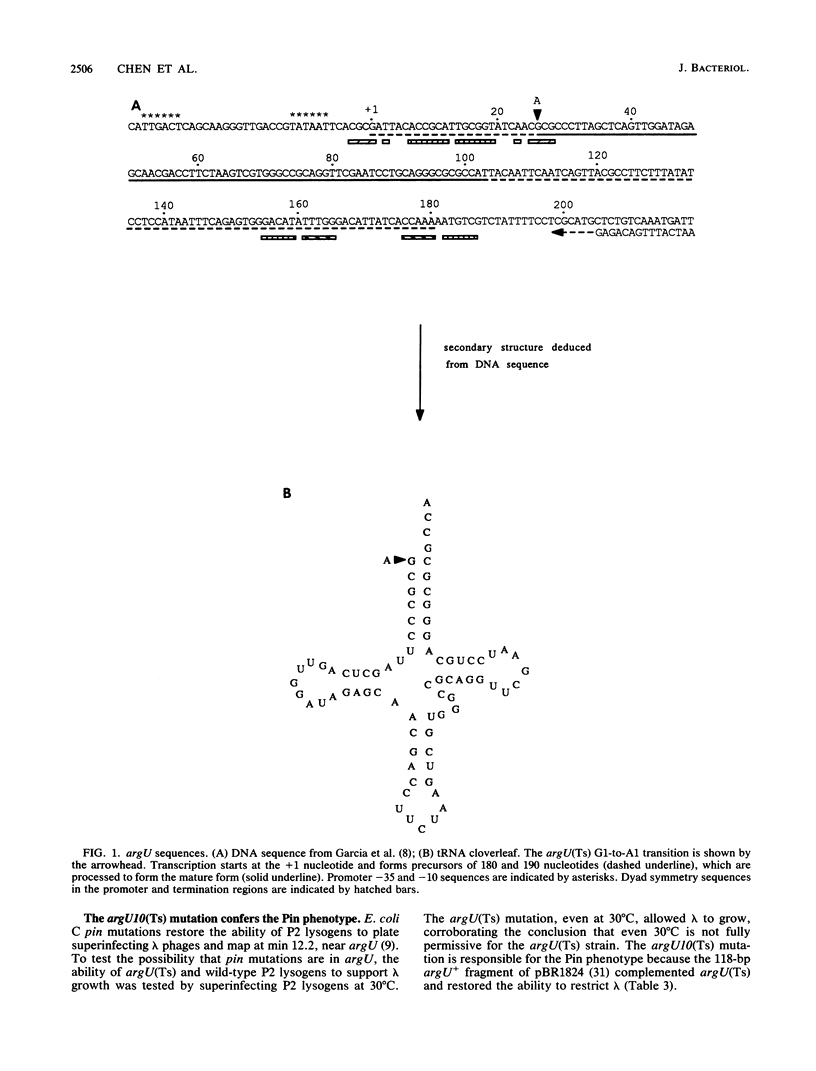
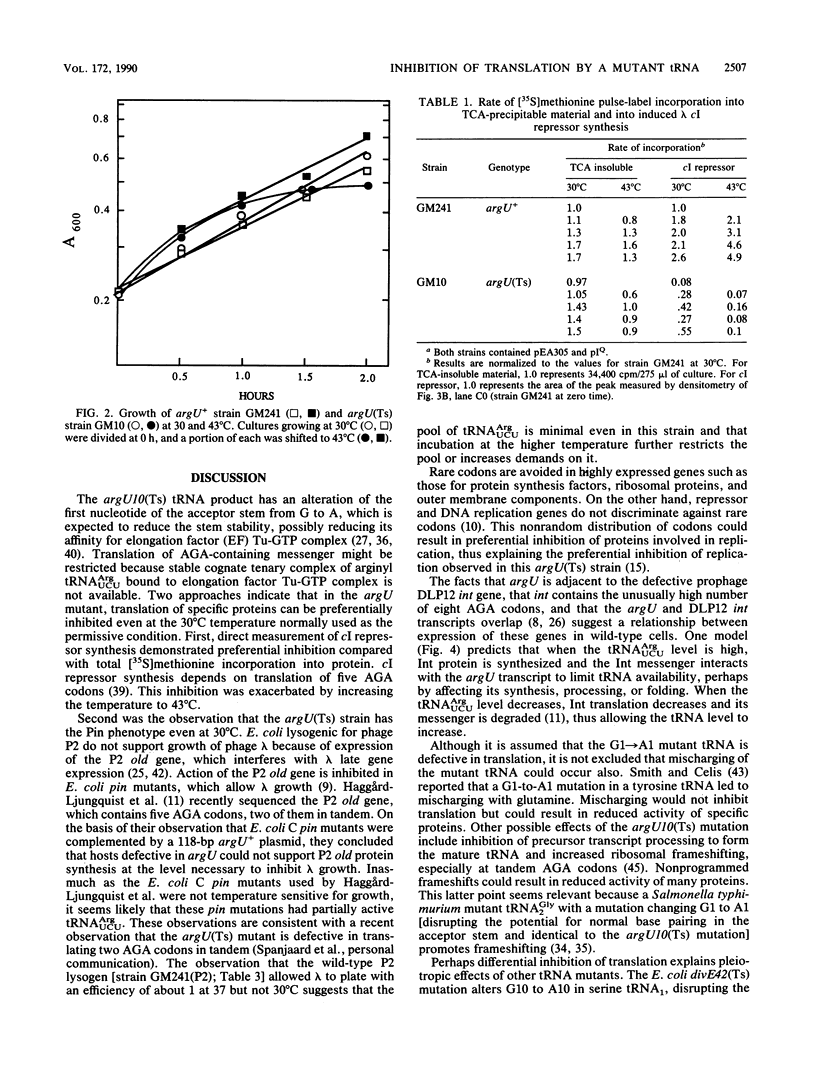
The Escherichia coli argU gene encodes a rare arginine tRNA (anticodon UCU) that translates the similarly rare AGA codon. The argU10(Ts) mutation is a transition that changes the first nucleotide of the mature tRNA from G to A, presumably destabilizing the acceptor stem. This mutation, when present in haploid condition in the chromosome, reduces the growth rate at 30 degrees C and results in cessation of growth after 60 to 90 min at 43 degrees C. The mutation also preferentially limits (compared with total protein synthesis) translation of an induced gene that depends on five AGA codons, i.e., the lambda cI repressor gene. Translation of another inducible protein, beta-galactosidase, which does not involve AGA codons, was inhibited to a much lesser extent. The chromosomal argU(Ts) mutation also confers the Pin phenotype, that is, loss of ability of the host, as a P2 lysogen, to inhibit growth of bacteriophage lambda, probably the result of reduced translation of the P2 old gene, which contains five AGA codons (E. Haggård-Ljungquist, V. Barreiro, R. Calendar, D. M. Kurnit, and H. Cheng, Gene 85:25-33, 1989).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Blum P., Holzschu D., Kwan H. S., Riggs D., Artz S. Gene replacement and retrieval with recombinant M13mp bacteriophages. J Bacteriol. 1989 Jan;171(1):538–546. doi: 10.1128/jb.171.1.538-546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis T. F., Maas W. K. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. IV. Further studies on the role of arginine transfer RNA repression of the enzymes of arginine biosynthesis. J Mol Biol. 1971 Nov 28;62(1):179–188. doi: 10.1016/0022-2836(71)90138-0. [DOI] [PubMed] [Google Scholar]
- Eggertsson G. Suppressors causing temperature sensitivity of growth in Escherichia coli. Genetics. 1968 Oct;60(2):269–280. doi: 10.1093/genetics/60.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M. J., Ozeki H. Structure and organization of the transfer ribonucleic acid genes of Escherichia coli K-12. Microbiol Rev. 1985 Dec;49(4):379–397. doi: 10.1128/mr.49.4.379-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. M., Mar P. K., Mullin D. A., Walker J. R., Prather N. E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986 May 9;45(3):453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- Ghisotti D., Zangrossi S., Sironi G. An Escherichia coli gene required for bacteriophage P2-lambda interference. J Virol. 1983 Dec;48(3):616–626. doi: 10.1128/jvi.48.3.616-626.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggård-Ljungquist E., Barreiro V., Calendar R., Kurnit D. M., Cheng H. The P2 phage old gene: sequence, transcription and translational control. Gene. 1989 Dec 21;85(1):25–33. doi: 10.1016/0378-1119(89)90460-5. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. M., Chu H., Irwin C. A., Walker J. R. Isolation and characterization of dnaX and dnaY temperature-sensitive mutants of Escherichia coli. Genetics. 1979 Aug;92(4):1041–1059. doi: 10.1093/genetics/92.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ishikura H., Nishimura S. Fractionation of serine transfer ribonucleic acids from Escherichia coli and their coding properties. Biochim Biophys Acta. 1968 Jan 29;155(1):72–81. doi: 10.1016/0005-2787(68)90336-5. [DOI] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor E. J., Baylis H. A., Chater K. F. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1987 Dec;1(10):1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- Leclerc G., Sirard C., Drapeau G. R. The Escherichia coli cell division mutation ftsM1 is in serU. J Bacteriol. 1989 Apr;171(4):2090–2095. doi: 10.1128/jb.171.4.2090-2095.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Kanda P., Kennedy R. C., Walker J. R. Relation of the Escherichia coli dnaX gene to its two products--the tau and gamma subunits of DNA polymerase III holoenzyme. Nucleic Acids Res. 1987 Oct 12;15(19):7663–7675. doi: 10.1093/nar/15.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Walker J. R. Escherichia coli DnaX product, the tau subunit of DNA polymerase III, is a multifunctional protein with single-stranded DNA-dependent ATPase activity. Proc Natl Acad Sci U S A. 1987 May;84(9):2713–2717. doi: 10.1073/pnas.84.9.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl G., Sironi G., Bialy H., Calendar R. Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc Natl Acad Sci U S A. 1970 Jul;66(3):587–594. doi: 10.1073/pnas.66.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey D. F., Mullin D. A., Walker J. R. Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J Bacteriol. 1989 Nov;171(11):6197–6205. doi: 10.1128/jb.171.11.6197-6205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A., Ribeiro N. S., Reid B. R., Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem. 1984 Apr 25;259(8):5010–5016. [PubMed] [Google Scholar]
- Mizusawa S., Ward D. F. A bacteriophage lambda vector for cloning with BamHI and Sau3A. Gene. 1982 Dec;20(3):317–322. doi: 10.1016/0378-1119(82)90200-1. [DOI] [PubMed] [Google Scholar]
- Mullin D. A., Garcia G. M., Walker J. R. An E. coli DNA fragment 118 base pairs in length provides dnaY+ complementing activity. Cell. 1984 Jun;37(2):669–674. doi: 10.1016/0092-8674(84)90399-4. [DOI] [PubMed] [Google Scholar]
- O'Mahony D. J., Hughes D., Thompson S., Atkins J. F. Suppression of a -1 frameshift mutation by a recessive tRNA suppressor which causes doublet decoding. J Bacteriol. 1989 Jul;171(7):3824–3830. doi: 10.1128/jb.171.7.3824-3830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony D. J., Mims B. H., Thompson S., Murgola E. J., Atkins J. F. Glycine tRNA mutants with normal anticodon loop size cause -1 frameshifting. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7979–7983. doi: 10.1073/pnas.86.20.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki M. Correlation between metabolism of phosphatidylglycerol and membrane synthesis in Escherichia coli. J Mol Biol. 1972 Jul 21;68(2):249–264. doi: 10.1016/0022-2836(72)90212-4. [DOI] [PubMed] [Google Scholar]
- Oki M., Mitsui H. Defective membrane synthesis in an E. coli mutant. Nature. 1974 Nov 1;252(5478):64–66. doi: 10.1038/252064a0. [DOI] [PubMed] [Google Scholar]
- Ono Y., Skoultchi A., Klein A., Lengyel P. Peptide chain elongation: discrimination against the initiator transfer RNA by microbial amino-acid polymerization factors. Nature. 1968 Dec 28;220(5174):1304–1307. doi: 10.1038/2201304a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Ohki M., Yura T., Ito K. Genetic studies of an Escherichia coli K-12 temperature-sensitive mutant defective in membrane protein synthesis. J Bacteriol. 1979 May;138(2):305–313. doi: 10.1128/jb.138.2.305-313.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T. DNA sequence of the bacteriophage gama cI gene. Nature. 1978 Nov 16;276(5685):301–302. doi: 10.1038/276301a0. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H., Sundari R. M. Structural requirements for recognition of Escherichia coli initiator and non-initiator transfer ribonucleic acids by bacterial T factor. J Biol Chem. 1974 Nov 25;249(22):7102–7110. [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Sironi G., Bialy H., Lozeron H. A., Calendar R. Bacteriophage P2: interaction with phage lambda and with recombination-deficient bacteria. Virology. 1971 Nov;46(2):387–396. doi: 10.1016/0042-6822(71)90040-7. [DOI] [PubMed] [Google Scholar]
- Spanjaard R. A., van Duin J. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura F., Nishimura S., Ohki M. The E. coli divE mutation, which differentially inhibits synthesis of certain proteins, is in tRNASer1. EMBO J. 1984 May;3(5):1103–1107. doi: 10.1002/j.1460-2075.1984.tb01936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjarnardóttir S., Uemura H., Dingermann T., Rafnar T., Thorsteinsdóttir S., Söll D., Eggertsson G. Escherichia coli supH suppressor: temperature-sensitive missense suppression caused by an anticodon change in tRNASer2. J Bacteriol. 1985 Jan;161(1):207–211. doi: 10.1128/jb.161.1.207-211.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 1984 Mar 26;12(6):2605–2618. doi: 10.1093/nar/12.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980 Feb;141(2):973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaizumi Z., Kuchino Y., Harada F., Nishimura S., McCloskey J. A. Primary structure of Escherichia coli tRNA UUR Leu. Presence of an unknown adenosine derivative in the first position of the anticodon which recognizes the UU codon series. J Biol Chem. 1980 Mar 10;255(5):2220–2225. [PubMed] [Google Scholar]



