Abstract
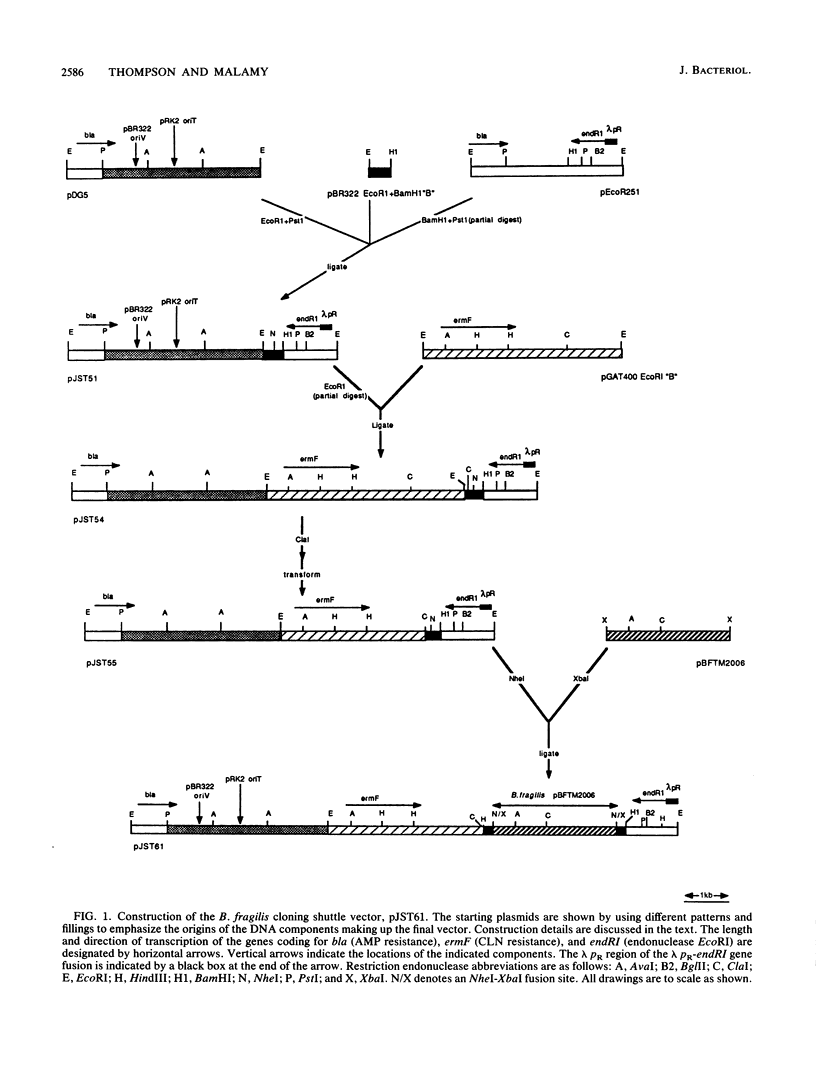
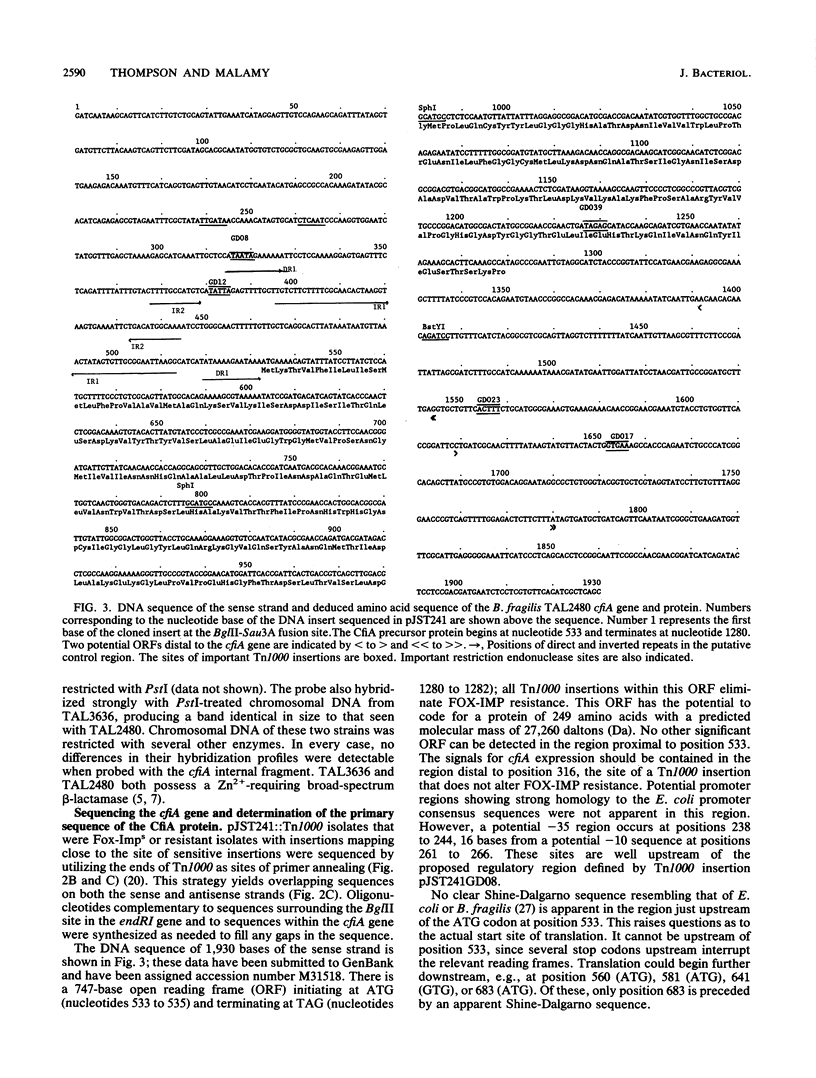
Using a newly constructed Bacteroides fragilis-Escherichia coli cloning shuttle vector, pJST61, we have cloned the cefoxitin (FOX)-imipenem (IMP) resistance determinant from B. fragilis TAL2480. FOX-IMP resistance in this strain results from the production of a periplasmic, Zn2(+)-containing beta-lactamase which hydrolyzes carbapenems and cephamycins and whose activity is resistant to clavulanic acid but sensitive to Zn2(+)-binding reagents, including EDTA. The pJST61 vector permits efficient library construction in E. coli and allows for the transfer of the library to B. fragilis recipients for the screening or selection of specific phenotypes. The library clone containing the FOX-IMP resistance gene was detected after transfer to B. fragilis TM4000 (Fox-Imps) selecting for Foxr. One of the isolates carrying plasmid pJST241 is resistant to FOX and IMP and synthesizes a periplasmic protein with substrate and inhibitor properties identical to those of strain TAL2480. On the basis of deletion analysis, Tn1000 insertion mutations, and DNA sequencing, we have defined the 747-base cfiA (FOX-IMP resistance) gene within the 3.6-kilobase cloned insert in pJST241. The cfiA gene contains an open reading frame that could code for a precursor protein of 249 amino acids and with a molecular mass of 27,260 daltons. A potential signal sequence has been identified at the N terminus of this protein; cleavage within this sequence would result in a protein of 231 amino acids with a molecular mass of 25,249 daltons. The CfiA protein shows remarkable similarities to the exported, Zn2(+)-requiring, type II beta-lactamase Blm proteins from Bacillus cereus 569/H and 5/B/6. Although overall amino acid identity is only 32%, the Zn ligand-binding His and Cys residues are precisely conserved and the amino acids in the vicinity of these sites show strong similarities (greater than 80%) when the CfiA and Blm proteins are compared.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Baldwin G. S., Waley S. G., Abraham E. P. Identification of histidine residues that act as zinc ligands in beta-lactamase II by differential tritium exchange. Biochem J. 1979 Jun 1;179(3):459–463. doi: 10.1042/bj1790459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Malamy M. H., Tally F. P. Beta-lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986 Nov;30(5):645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Tally F. P., Jacobus N. V., Aldridge K., Cleary T., Finegold S. M., Hill G., Iannini P., O'Keefe J. P., Pierson C. Susceptibility of the Bacteroides fragilis group in the United States: analysis by site of isolation. Antimicrob Agents Chemother. 1988 May;32(5):717–722. doi: 10.1128/aac.32.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Tally F. P., Jacobus N. V., Marsh P. K., Mayhew J. W. Cefoxitin inactivation by Bacteroides fragilis. Antimicrob Agents Chemother. 1983 Dec;24(6):936–940. doi: 10.1128/aac.24.6.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Tally F. P., Storey J. R., Malamy M. H. Transfer of beta-lactamase-associated cefoxitin resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986 May;29(5):918–920. doi: 10.1128/aac.29.5.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne J., Vézina G., Levesque R. C. Cloning and expression of the imipenem-hydrolyzing beta-lactamase operon from Pseudomonas maltophilia in Escherichia coli. Antimicrob Agents Chemother. 1988 Jun;32(6):819–826. doi: 10.1128/aac.32.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Bouic K., Matthews B. Genetic transfer systems in Bacteroides: cloning and mapping of the transferable tetracycline-resistance locus. Mol Microbiol. 1989 Nov;3(11):1617–1623. doi: 10.1111/j.1365-2958.1989.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Davis C. E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Hecht D. W., Malamy M. H. Tn4399, a conjugal mobilizing transposon of Bacteroides fragilis. J Bacteriol. 1989 Jul;171(7):3603–3608. doi: 10.1128/jb.171.7.3603-3608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill H. A., Sammes P. G., Waley S. G. Active sites of beta-lactamases from Bacillus cereus. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):333–344. doi: 10.1098/rstb.1980.0050. [DOI] [PubMed] [Google Scholar]
- Hurlbut S., Cuchural G. J., Tally F. P. Imipenem resistance in Bacteroides distasonis mediated by a novel beta-lactamase. Antimicrob Agents Chemother. 1990 Jan;34(1):117–120. doi: 10.1128/aac.34.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Carlino A., Madonna M. J., Lampen J. O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985 Oct;164(1):223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T., Williams J. D. beta-Lactamases of subspecies of Bacteroides fragilis. J Antimicrob Chemother. 1978 Jul;4(B):47–54. doi: 10.1093/jac/4.suppl_b.47. [DOI] [PubMed] [Google Scholar]
- Lim H. M., Pène J. J., Shaw R. W. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 beta-lactamase II structural gene. J Bacteriol. 1988 Jun;170(6):2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Whalen W., Das A., Berg C. M. Rapid sequencing of cloned DNA using a transposon for bidirectional priming: sequence of the Escherichia coli K-12 avtA gene. Nucleic Acids Res. 1987 Nov 25;15(22):9461–9469. doi: 10.1093/nar/15.22.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. K., Malamy M. H., Shimell M. J., Tally F. P. Sequence homology of clindamycin resistance determinants in clinical isolates of Bacteroides spp. Antimicrob Agents Chemother. 1983 May;23(5):726–730. doi: 10.1128/aac.23.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Lanka E., Malamy M. H. F factor inhibition of conjugal transfer of broad-host-range plasmid RP4: requirement for the protein product of pif operon regulatory gene pifC. J Bacteriol. 1985 Sep;163(3):1067–1073. doi: 10.1128/jb.163.3.1067-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson B., Dornbusch K., Nord C. E. Factors contributing to resistance to beta-lactam antibiotics in Bacteroides fragilis. Antimicrob Agents Chemother. 1979 Feb;15(2):263–268. doi: 10.1128/aac.15.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J. L., Odelson D. A., Macrina F. L. Complete nucleotide sequence and transcription of ermF, a macrolide-lincosamide-streptogramin B resistance determinant from Bacteroides fragilis. J Bacteriol. 1986 Nov;168(2):523–533. doi: 10.1128/jb.168.2.523-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robillard N. J., Tally F. P., Malamy M. H. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J Bacteriol. 1985 Dec;164(3):1248–1255. doi: 10.1128/jb.164.3.1248-1255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino Y., Inoue M., Mitsuhashi S. Purification and properties of an inducible cephalosporinase from Pseudomonas maltophilia GN12873. Antimicrob Agents Chemother. 1984 Mar;25(3):362–365. doi: 10.1128/aac.25.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Fujii T., Okamoto R., Inoue M., Mitsuhashi S. Biochemical properties of beta-lactamase produced by Flavobacterium odoratum. Antimicrob Agents Chemother. 1985 Apr;27(4):612–614. doi: 10.1128/aac.27.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Barber R. D., Salyers A. A. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J Bacteriol. 1989 Mar;171(3):1294–1302. doi: 10.1128/jb.171.3.1294-1302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern J. A., Parker J. R., Woods D. R. Expression and purification of glutamine synthetase cloned from Bacteroides fragilis. J Gen Microbiol. 1986 Oct;132(10):2827–2835. doi: 10.1099/00221287-132-10-2827. [DOI] [PubMed] [Google Scholar]
- Strauch K. L., Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tally F. P., Cuchural G. J., Jr, Jacobus N. V., Gorbach S. L., Aldridge K., Cleary T., Finegold S. M., Hill G., Iannini P., O'Keefe J. P. Nationwide study of the susceptibility of the Bacteroides fragilis group in the United States. Antimicrob Agents Chemother. 1985 Nov;28(5):675–677. doi: 10.1128/aac.28.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tally F. P., Snydman D. R., Gorbach S. L., Malamy M. H. Plasmid-mediated, transferable resistance to clindamycin and erythromycin in Bacteroides fragilis. J Infect Dis. 1979 Jan;139(1):83–88. doi: 10.1093/infdis/139.1.83. [DOI] [PubMed] [Google Scholar]


