Abstract
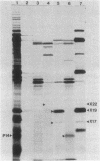
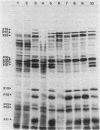
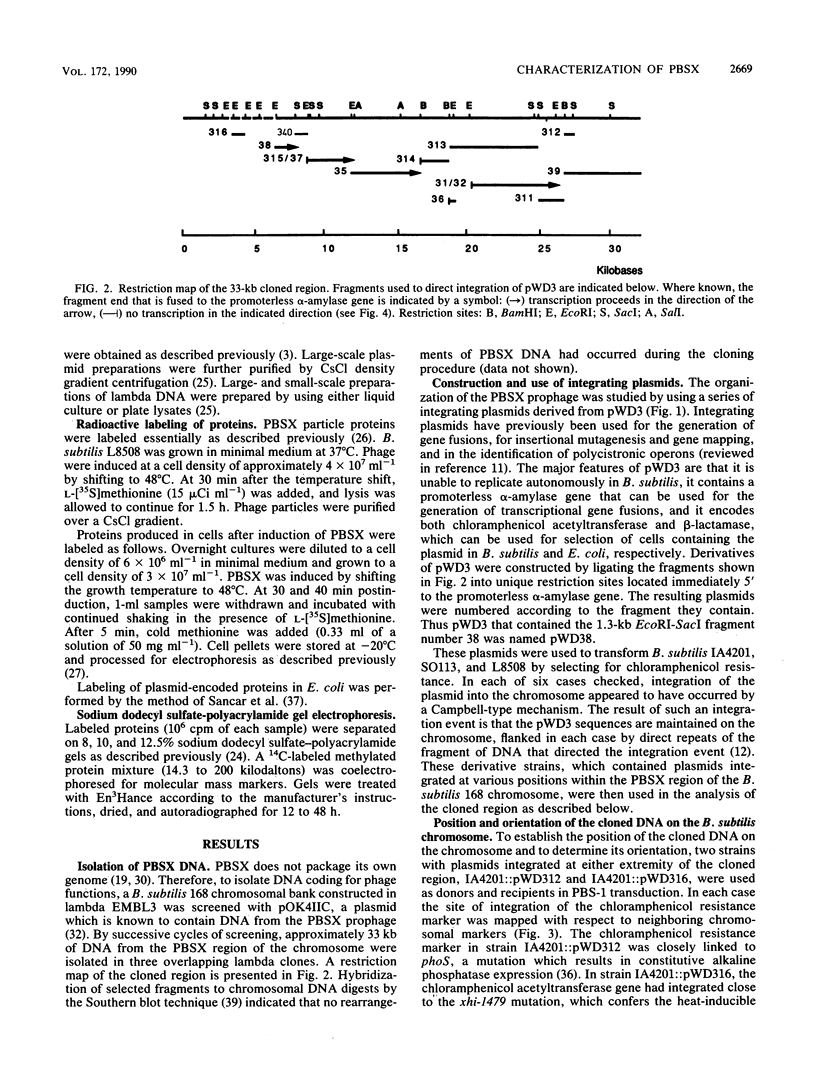
PBSX, a defective Bacillus subtilis prophage, maps to the metA-metC region of the chromosome. DNA (33 kilobases) from this region of the chromosome was cloned and analyzed by insertional mutagenesis with the integrating plasmid pWD3. This plasmid had a promoterless alpha-amylase gene (amyL) that provided information on the direction and level of transcription at the site of integration. Transcription under the control of the PBSX repressor proceeded in the direction metA to metC over a distance of at least 18 kilobases. Electrophoretic analysis of proteins produced by different integrant strains upon PBSX induction and by fragments subcloned in Escherichia coli allowed the identification of early and late regions of the prophage. A set of contiguous fragments directing mutagenic integration suggested that the minimum size of an operon that encodes phage structural proteins is 19 kilobases. The adaptation of PBSX transcriptional and replicational functions to a chromosomally based, thermoinducible expression system is discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson L. M., Bott K. F. DNA packaging by the Bacillus subtilis defective bacteriophage PBSX. J Virol. 1985 Jun;54(3):773–780. doi: 10.1128/jvi.54.3.773-780.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. M., Ruley H. E., Bott K. F. Isolation of an autonomously replicating DNA fragment from the region of defective bacteriophage PBSX of Bacillus subtilis. J Bacteriol. 1982 Jun;150(3):1280–1286. doi: 10.1128/jb.150.3.1280-1286.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967 Dec;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton R. S. Prophage mutation causing heat inducibility of defective Bacillus subtilis bacteriophage PBSX. J Virol. 1976 Oct;20(1):22–28. doi: 10.1128/jvi.20.1.22-28.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton R. S. Selection of Bacillus subtilis 168 mutants with deletions of the PBSX prophage. J Gen Virol. 1980 Feb;46(2):427–437. doi: 10.1099/0022-1317-46-2-427. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- Dhaese P., Hussey C., Van Montagu M. Thermo-inducible gene expression in Bacillus subtilis using transcriptional regulatory elements from temperate phage phi 105. Gene. 1984 Dec;32(1-2):181–194. doi: 10.1016/0378-1119(84)90046-5. [DOI] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Mechanism of integrating foreign DNA during transformation of Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3664–3668. doi: 10.1073/pnas.75.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Garro A. J., Leffert H., Marmur J. Genetic mapping of a defective bacteriophage on the chromosome of Bacillus subtilis 168. J Virol. 1970 Sep;6(3):340–343. doi: 10.1128/jvi.6.3.340-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro A. J., Marmur J. Defective bacteriophages. J Cell Physiol. 1970 Dec;76(3):253–263. doi: 10.1002/jcp.1040760305. [DOI] [PubMed] [Google Scholar]
- Gryczan T., Contente S., Dubnau D. Molecular cloning of heterologous chromosomal DNA by recombination between a plasmid vector and a homologous resident plasmid in Bacillus subtilis. Mol Gen Genet. 1980 Feb;177(3):459–467. doi: 10.1007/BF00271485. [DOI] [PubMed] [Google Scholar]
- Haas M., Yoshikawa H. Defective bacteriophage PBSH in Bacillus subtilis. II. Intracellular development of the induced prophage. J Virol. 1969 Feb;3(2):248–260. doi: 10.1128/jvi.3.2.248-260.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Marmur J. Characterization of inducible bacteriophages in Bacillus licheniformis. J Virol. 1970 Feb;5(2):237–246. doi: 10.1128/jvi.5.2.237-246.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Niaudet B., Pierre E., Ehrlich S. D. Stable gene amplification in the chromosome of Bacillus subtilis. Gene. 1985;40(1):47–55. doi: 10.1016/0378-1119(85)90023-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mauël C., Karamata D. Characterization of proteins induced by mitomycin C treatment of Bacillus subtilis. J Virol. 1984 Mar;49(3):806–812. doi: 10.1128/jvi.49.3.806-812.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P., Dowds B. C., McConnell D. J., Devine K. M. Oxidative stress and growth temperature in Bacillus subtilis. J Bacteriol. 1987 Dec;169(12):5766–5770. doi: 10.1128/jb.169.12.5766-5770.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Goze A., Ehrlich S. D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982 Oct;19(3):277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- O'Kane C., Stephens M. A., McConnell D. Integrable alpha-amylase plasmid for generating random transcriptional fusions in Bacillus subtilis. J Bacteriol. 1986 Nov;168(2):973–981. doi: 10.1128/jb.168.2.973-981.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Mudd J. A., Mangan J., Huang W. M., Subbaiah T. V., Marmur J. Properties of the defective phage of Bacillus subtilis. J Mol Biol. 1968 Jun 28;34(3):413–428. doi: 10.1016/0022-2836(68)90169-1. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Mudd J. A., Marmur J. Conversion of Bacillus subtilis DNA to phage DNA following mitomycin C induction. J Mol Biol. 1968 Jun 28;34(3):429–437. doi: 10.1016/0022-2836(68)90170-8. [DOI] [PubMed] [Google Scholar]
- Ortlepp S. A., Ollington J. F., McConnell D. J. Molecular cloning in Bacillus subtilis of a Bacillus licheniformis gene encoding a thermostable alpha amylase. Gene. 1983 Sep;23(3):267–276. doi: 10.1016/0378-1119(83)90017-3. [DOI] [PubMed] [Google Scholar]
- Osburne M. S., Craig R. J., Rothstein D. M. Thermoinducible transcription system for Bacillus subtilis that utilizes control elements from temperate phage phi 105. J Bacteriol. 1985 Sep;163(3):1101–1108. doi: 10.1128/jb.163.3.1101-1108.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Curtis C. A., de Lencastre H. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Taylor S. Y. New types of mutation affecting formation of alkaline phosphatase by Bacillus subtilis in sporulation conditions. J Gen Microbiol. 1977 Sep;102(1):69–80. doi: 10.1099/00221287-102-1-69. [DOI] [PubMed] [Google Scholar]
- SEAMAN E., TARMY E., MARMUR J. INDUCIBLE PHAGES OF BACILLUS SUBTILIS. Biochemistry. 1964 May;3:607–613. doi: 10.1021/bi00893a001. [DOI] [PubMed] [Google Scholar]
- STICKLER D. J., TUCKER R. G., KAY D. BACTERIOPHAGE-LIKE PARTICLES RELEASED FROM BACILLUS SUBTILIS AFTER INDUCTION WITH HYDROGEN PEROXIDE. Virology. 1965 May;26:142–145. doi: 10.1016/0042-6822(65)90035-8. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steensma H. Y., Robertson L. A., van Elsas J. D. The occurrence and taxonomic value of PBS X-like defective phages in the genus Bacillus. Antonie Van Leeuwenhoek. 1978;44(3-4):353–366. doi: 10.1007/BF00394312. [DOI] [PubMed] [Google Scholar]
- Tagg J. R., Dajani A. S., Wannamaker L. W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976 Sep;40(3):722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurm P., Garro A. J. Bacteriophage-specific protein synthesis during induction of the defective Bacillus subtilis bacteriophage PBSX. J Virol. 1975 Jul;16(1):179–183. doi: 10.1128/jvi.16.1.179-183.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurm P., Garro A. J. Isolation and characterization of prophage mutants of the defective Bacillus subtilis bacteriophage PBSX. J Virol. 1975 Jul;16(1):184–191. doi: 10.1128/jvi.16.1.184-191.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]