Abstract
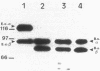
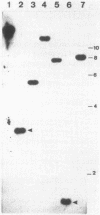
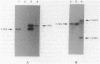
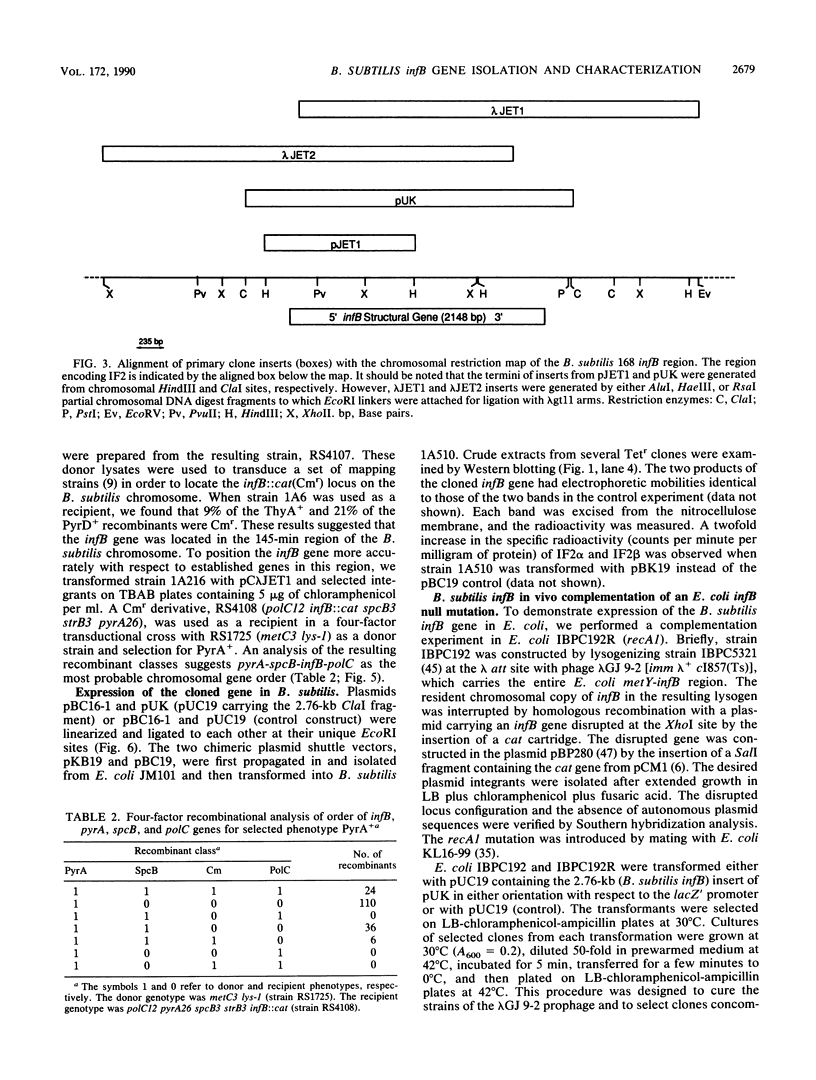
Western blot (immunoblot) analysis of Bacillus subtilis cell extracts detected two proteins that cross-reacted with monospecific polyclonal antibody raised against Escherichia coli initiation factor 2 alpha (IF2 alpha). Subsequent Southern blot analysis of B. subtilis genomic DNA identified a 1.3-kilobase (kb) HindIII fragment which cross-hybridized with both E. coli and Bacillus stearothermophilus IF2 gene probes. This DNA was cloned from a size-selected B. subtilis plasmid library. The cloned HindIII fragment, which was shown by DNA sequence analysis to encode the N-terminal half of the B. subtilis IF2 protein and 0.2 kb of upstream flanking sequence, was utilized as a homologous probe to clone an overlapping 2.76-kb ClaI chromosomal fragment containing the entire IF2 structural gene. The HindIII fragment was also used as a probe to obtain overlapping clones from a lambda gt11 library which contained additional upstream and downstream flanking sequences. Sequence comparisons between the B. subtilis IF2 gene and the other bacterial homologs from E. coli, B. stearothermophilus, and Streptococcus faecium displayed extensive nucleic acid and protein sequence homologies. The B. subtilis infB gene encodes two proteins, IF2 alpha (78.6 kilodaltons) and IF2 beta (68.2 kilodaltons); both were expressed in B. subtilis and E. coli. These two proteins cross-reacted with antiserum to E. coli IF2 alpha and were able to complement in vivo an E. coli infB gene disruption. Four-factor recombination analysis positioned the infB gene at 145 degrees on the B. subtilis chromosome, between the polC and spcB loci. This location is distinct from those of the other major ribosomal protein and rRNA gene clusters of B. subtilis.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brombach M., Gualerzi C. O., Nakamura Y., Pon C. L. Molecular cloning and sequence of the Bacillus stearothermophilus translational initiation factor IF2 gene. Mol Gen Genet. 1986 Oct;205(1):97–102. doi: 10.1007/BF02428037. [DOI] [PubMed] [Google Scholar]
- Cenatiempo Y., Deville F., Brot N., Weissbach H. In vitro expression of the Escherichia coli nusA-infB operon. J Biol Chem. 1987 Jan 5;262(1):152–157. [PubMed] [Google Scholar]
- Cenatiempo Y., Deville F., Dondon J., Grunberg-Manago M., Sacerdot C., Hershey J. W., Hansen H. F., Petersen H. U., Clark B. F., Kjeldgaard M. The protein synthesis initiation factor 2 G-domain. Study of a functionally active C-terminal 65-kilodalton fragment of IF2 from Escherichia coli. Biochemistry. 1987 Aug 11;26(16):5070–5076. doi: 10.1021/bi00390a028. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Cole J. R., Olsson C. L., Hershey J. W., Grunberg-Manago M., Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J Mol Biol. 1987 Dec 5;198(3):383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- Coppard N. J., Cramer F., Clark B. F. Identification of elongation factor 1 alpha from mouse liver. FEBS Lett. 1982 Aug 23;145(2):332–336. doi: 10.1016/0014-5793(82)80194-4. [DOI] [PubMed] [Google Scholar]
- Dedonder R. A., Lepesant J. A., Lepesant-Kejzlarová J., Billault A., Steinmetz M., Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol. 1977 Apr;33(4):989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondon J., Plumbridge J. A., Hershey J. W., Grunberg-Manago M. Overproduction and purification of initiation factor IF2 and pNUSA proteins from a recombinant plasmid bearing strain. Biochimie. 1985 Jun;67(6):643–649. doi: 10.1016/s0300-9084(85)80206-6. [DOI] [PubMed] [Google Scholar]
- Edwards H., Schimmel P. An E. coli aminoacyl-tRNA synthetase can substitute for yeast mitochondrial enzyme function in vivo. Cell. 1987 Nov 20;51(4):643–649. doi: 10.1016/0092-8674(87)90133-4. [DOI] [PubMed] [Google Scholar]
- Friedrich K., Brombach M., Pon C. L. Identification, cloning and sequence of the Streptococcus faecium infB (translational initiation factor IF2) gene. Mol Gen Genet. 1988 Nov;214(3):595–600. doi: 10.1007/BF00330501. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989 Dec;3(12A):1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. A sensitive immunoblotting method for measuring protein synthesis initiation factor levels in lysates of Escherichia coli. J Biol Chem. 1981 Dec 25;256(24):12836–12839. [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. The rate of evolutionary divergence of initiation factors IF2 and IF3 in various bacterial species determined quantitatively by immunoblotting. Arch Microbiol. 1984 Dec;140(2-3):187–192. doi: 10.1007/BF00454924. [DOI] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Kreft J., Hughes C. Cloning vectors derived from plasmids and phage of Bacillus. Curr Top Microbiol Immunol. 1982;96:1–17. doi: 10.1007/978-3-642-68315-2_1. [DOI] [PubMed] [Google Scholar]
- Kurihara T., Nakamura Y. Cloning of the nusA gene of Escherichia coli. Mol Gen Genet. 1983;190(2):189–195. doi: 10.1007/BF00330639. [DOI] [PubMed] [Google Scholar]
- Köhler T., Harayama S., Ramos J. L., Timmis K. N. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J Bacteriol. 1989 Aug;171(8):4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leighton T. J., Doi R. H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971 May 25;246(10):3189–3195. [PubMed] [Google Scholar]
- Leighton T. Sporulation-specific translational discrimination in Bacillus subtilis. J Mol Biol. 1974 Jul 15;86(4):855–863. doi: 10.1016/0022-2836(74)90358-1. [DOI] [PubMed] [Google Scholar]
- Love E., D'Ambrosio D., Brown N. C. Mapping of the gene specifying DNA polymerase III of Bacillus subtilis. Mol Gen Genet. 1976 Mar 30;144(3):313–321. doi: 10.1007/BF00341730. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López P., Martinez S., Diaz A., Espinosa M. Streptococcus pneumoniae polA gene is expressed in Escherichia coli and can functionally substitute for the E. coli polA gene. J Bacteriol. 1987 Oct;169(10):4869–4871. doi: 10.1128/jb.169.10.4869-4871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez S., Lopez P., Espinosa M., Lacks S. A. Complementation of Bacillus subtilis polA mutants by DNA polymerase I from Streptococcus pneumoniae. Mol Gen Genet. 1987 Dec;210(2):203–210. doi: 10.1007/BF00325685. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Mizusawa S. In vivo evidence that the nusA and infB genes of E. coli are part of the same multi-gene operon which encodes at least four proteins. EMBO J. 1985 Feb;4(2):527–532. doi: 10.1002/j.1460-2075.1985.tb03660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Plumbridge J., Dondon J., Grunberg-Manago M. Evidence for autoregulation of the nusA-infB operon of Escherichia coli. Gene. 1985;36(1-2):189–193. doi: 10.1016/0378-1119(85)90085-x. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Munoz L., Doi R. H. A procedure to remove protease activities from Bacillus subtilis sporulating cells and their crude extracts. Anal Biochem. 1977 Mar;78(1):165–170. doi: 10.1016/0003-2697(77)90020-3. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A., Deville F., Sacerdot C., Petersen H. U., Cenatiempo Y., Cozzone A., Grunberg-Manago M., Hershey J. W. Two translational initiation sites in the infB gene are used to express initiation factor IF2 alpha and IF2 beta in Escherichia coli. EMBO J. 1985 Jan;4(1):223–229. doi: 10.1002/j.1460-2075.1985.tb02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A., Dondon J., Nakamura Y., Grunberg-Manago M. Effect of NusA protein on expression of the nusA,infB operon in E. coli. Nucleic Acids Res. 1985 May 10;13(9):3371–3388. doi: 10.1093/nar/13.9.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A., Howe J. G., Springer M., Touati-Schwartz D., Hershey J. W., Grunberg-Manago M. Cloning and mapping of a gene for translational initiation factor IF2 in Escherichia coli. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5033–5037. doi: 10.1073/pnas.79.16.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Migot C., Grumberg-Manago M. Cloning of E. coli pnp gene from an episome. Mol Gen Genet. 1981;183(2):298–305. doi: 10.1007/BF00270632. [DOI] [PubMed] [Google Scholar]
- Price C. W., Doi R. H. Genetic mapping of rpoD implicates the major sigma factor of Bacillus subtilis RNA polymerase in sporulation initiation. Mol Gen Genet. 1985;201(1):88–95. doi: 10.1007/BF00397991. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sacerdot C., Dessen P., Hershey J. W., Plumbridge J. A., Grunberg-Manago M. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7787–7791. doi: 10.1073/pnas.81.24.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands J. F., Cummings H. S., Sacerdot C., Dondon L., Grunberg-Manago M., Hershey J. W. Cloning and mapping of infA, the gene for protein synthesis initiation factor IF1. Nucleic Acids Res. 1987 Jul 10;15(13):5157–5168. doi: 10.1093/nar/15.13.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands J. F., Regnier P., Cummings H. S., Grunberg-Manago M., Hershey J. W. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 1988 Nov 25;16(22):10803–10816. doi: 10.1093/nar/16.22.10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R. A., Leighton T. Intergenic suppressors of temperature-sensitive sporulation in Bacillus subtilis are allele non-specific. Mol Gen Genet. 1981;183(3):532–537. doi: 10.1007/BF00268777. [DOI] [PubMed] [Google Scholar]
- Sharrock R. A., Leighton T., Wittmann H. G. Macrolide and aminoglycoside antibiotic resistance mutations in the bacillus subtilis ribosome resulting in temperature-sensitive sporulation. Mol Gen Genet. 1981;183(3):538–543. doi: 10.1007/BF00268778. [DOI] [PubMed] [Google Scholar]
- Sobel E., Martinez H. M. A multiple sequence alignment program. Nucleic Acids Res. 1986 Jan 10;14(1):363–374. doi: 10.1093/nar/14.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Graffe M., Grunberg-Manago M. Characterization of an E. coli mutant with a thermolabile initiation factor IF3 activity. Mol Gen Genet. 1977 Feb 28;151(1):17–26. doi: 10.1007/BF00446908. [DOI] [PubMed] [Google Scholar]
- Springer M., Graffe M., Grunberg-Manago M. Genetic organization of the E. coli chromosome around the structural gene for initiation factor IF3 (infC). Mol Gen Genet. 1979 Feb 1;169(3):337–343. doi: 10.1007/BF00382279. [DOI] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J. W., Boylan S. A., Price C. W. Gene for the alpha subunit of Bacillus subtilis RNA polymerase maps in the ribosomal protein gene cluster. J Bacteriol. 1986 Oct;168(1):65–71. doi: 10.1128/jb.168.1.65-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiboni O., Di Pasquale G. Organization of genes for ribosomal proteins S7 and S12, elongation factors EF-Tu and EF-G in the cyanobacterium Spirulina platensis. Biochim Biophys Acta. 1987 Feb 27;908(2):113–122. doi: 10.1016/0167-4781(87)90050-9. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Johnson C. W., Ginther C. L., Leighton T., Wittmann H. G. Erythromycin resistant mutations in Bacillus subtilis cause temperature sensitive sporulation. Mol Gen Genet. 1977 Jan 18;150(2):147–159. doi: 10.1007/BF00695395. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]