Abstract
While greater than 80% of angiotensin II (Ang II) formation in the human heart and greater than 60% in arteries appears to result from chymase activity, no cardiovascular cell–expressed chymase has been previously reported. We now describe the cloning of a full-length cDNA encoding a novel chymase from rat vascular smooth muscle cells. The cDNA encompasses 953 nucleotides, encodes 247 amino acids, and exhibits 74% and 80% homology in amino acid sequence to rat mast cell chymase I and II, respectively. Southern blot analysis indicates that the rat vascular chymase is encoded by a separate gene. This chymase was induced in hypertrophied rat pulmonary arteries, with 11-fold and 8-fold higher chymase mRNA levels in aortic and pulmonary artery smooth muscle cells from spontaneously hypertensive than in corresponding tissues from normotensive rats. We assayed the activity of the endogenous enzyme and of a recombinant, epitope-tagged chymase in transfected smooth muscle cells and showed that Ang II production from Ang I can be inhibited with chymostatin, but not EDTA or captopril. Spontaneously hypertensive rats show elevated chymase expression and increased chymostatin-inhibitable angiotensin-converting activity, suggesting a possible role for this novel enzyme in the pathophysiology of hypertension.
Introduction
The renin-angiotensin system regulates blood pressure and is involved in remodeling of both the heart and blood vessels in pulmonary (1, 2) and systemic hypertension and atherosclerosis (3–5). The proposed mechanisms of angiotensin II–induced (Ang II-induced) vascular remodeling have been addressed in cultured cells. Ang II enhances type I collagen synthesis in cardiovascular fibroblasts (6) and induces myocyte and vascular smooth muscle cell (SMC) proliferation and hypertrophy (7, 8). In clinical studies, blockade of the renin-angiotensin system with angiotensin-converting enzyme (ACE) inhibitors has been of therapeutic benefit in hypertension and heart failure (9, 10). Experimental studies with these agents have shown significant regression of left ventricular hypertrophy (11) and atherosclerosis (12, 13). However, the observation that ACE inhibitors incompletely block Ang II formation led to the discovery that other enzymes might produce Ang II both in the heart and blood vessels (14, 15).
In the human heart, only 10–20% of Ang II formation can be attributed to ACE activity (16). Non–ACE-dependent Ang II formation was also reported in the heart of the dog (15), hamster (17), and baboon (18). A human heart chymase cDNA was cloned, and the enzyme was purified and characterized as a chymase because it was inhibited by chymostatin and had the property of producing Ang II from Ang I (19, 20). A polyclonal Ab localized this enzyme to the interstitium of the myocardium, specifically, the granules of mast cells, endothelial cells, and cells described as “mesenchymal,” but not myocytes (21). Human chymase was later found by Western immunoblot analysis to be widely distributed in other tissues, e.g., uterus, tonsil, stomach, esophagus, coronary artery, and aorta (22), but its cellular origin in these tissues was not determined.
A chymostatin-sensitive Ang II–generating enzyme has been reported in the human, monkey, dog, and hamster vasculature (23–26). The enzyme was purified from human gastroepiploic arteries and hamster cheek-pouch vessels. In the human arteries, NH2-terminal amino acid–sequence analysis suggested that it was the same mast cell chymase described in human heart/skin (27, 28). Increased mRNA expression of this chymase was found in the monkey atherosclerotic aorta (29). Immunostaining of human coronary and carotid atherosclerotic lesions with mAb’s, however, localized this chymase to mast cells infiltrating these lesions (30). Since the blood vessel wall is the site of Ang II generation, one might expect a chymase to be locally produced. But to date, no specific chymase expressed or synthesized by arterial endothelial or SMCs had been identified. Isolation and identification of such an enzyme could be important as a potential therapeutic target.
Although in rodents Ang II formation is primarily ACE-related, non–ACE-dependent Ang II formation is also found in rat heart (31, 32) and vasculature (33–35) and in cultured vascular cells (36, 37). We now report the cloning and sequencing of a full-length cDNA encoding a novel rat vascular chymase (RVCH) from pulmonary artery SMCs. In situ hybridization also revealed increased RVCH expression in arteries from rats with pulmonary hypertension. We further document chymase activity by fluorescent substrate assay as well as Ang II generation in A10 cell extracts and show that both functions are inhibited by chymostatin. Moreover, stable transfection of the RVCH cDNA in A10 cells results in a more than eightfold increase in chymase activity. Recombinant epitope tag from influenza hemagglutinin RVCH cDNA, when transfected in A10 cells, produces a chymase with chymostatin-inhibitable Ang II–generating properties and a localization pattern similar to that observed with endogenous chymase. There is also increased expression of RVCH mRNA in vascular SMCs from spontaneously hypertensive rats (SHR), compared with normotensive rats, which correlates with enhanced chymase protein and chymostatin-inhibitable angiotensin-converting activity. Taken together, these studies provide both structural and functional evidence that we have cloned a novel vascular chymase that may be relevant to the pathogenesis of hypertension.
Methods
Cloning of RVCH cDNA.
Pulmonary artery SMCs were obtained by explant technique from Sprague-Dawley rats (Charles River Laboratories, Wilmington, Massachusetts, USA) and characterized both by morphology and by a mAb that recognizes smooth muscle α-actin. RNA was extracted from SMCs using Trizol (Life Technologies Inc., Gaithersburg, Maryland, USA), according to the manufacturer’s instructions. Then 3′-RACE was used to clone and sequence cDNAs encoding novel serine proteases (38). The first-strand cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase and primer (dT)17-adaptor (5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′). A degenerate oligonucleotide primer, 5′-GGNGAYTCNGGNGGNCCNCT-3′ (Y = C or T; N = A, C, G, or T), was designed according to the amino acid sequence GDSGGP, which is conserved in serine proteases. An adaptor primer (5′-GACTCGAGTCGACATCGA-3′) was used for PCR. PCR consisted of a 4-minute denaturation at 95°C, followed by 35 cycles of 45 seconds at 95°C, 45 seconds at 55°C, and 30 seconds at 72°C. The fragments between 300 and 500 bp were isolated by gel purification and subcloned into pCR2.1 vector (Invitrogen Corp., Carlsbad, California, USA). Ninety colonies with an insert between 300 and 450 bp were sequenced. A sequence of 311 bp showing 84% homology to rat mast cell protease II (RMCP II) was identified, and the 5′ end of the cDNA was then obtained using 5′-RACE. Primers CD (5′-TATAAGACCTCAGGCTTGGAG-3′) and PC (5′-TCAGCTACTTTCCCTTAAGAC-3′) were designed according to the sequences obtained from 3′-RACE for cDNA synthesis and PCR, respectively (Figure 1a). The first cDNA strand was synthesized using primer CD and purified using QIAquick spin column. A poly(A) tail was added to the 3′ end of the cDNA using terminal deoxynucleotidyl-transferase. PCR was performed under the same conditions as described above. PCR products were subcloned into pCR2.1 vector. Two colonies with an insert of approximately 800 bp in length were isolated and sequenced. They were found to be identical and contained a region of 135-bp overlap with the sequence obtained by 3′-RACE. The full-length cDNA was obtained by RT-PCR. Two primers (forward primer: 5′-GAGAAGCTCACCCCAGGCTGCA-3′; reverse primer: 5′-GGAAATTATGTCTTTATTGAGG-3′) were used. The PCR products were subcloned and three colonies were isolated and sequenced.
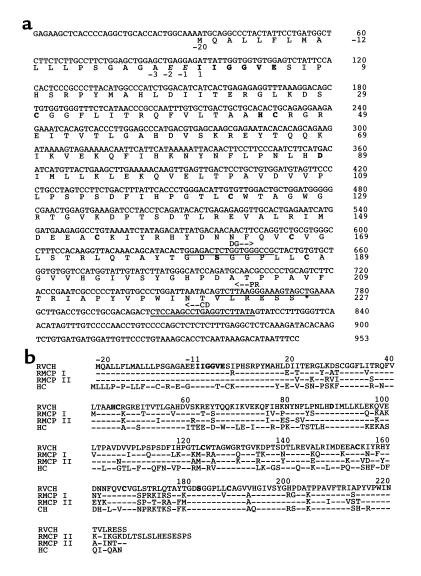
Figure 1.
Nucleotide sequence and deduced amino acid sequence of RVCH. (a) The nucleotide sequences annealing to the degenerate primer (DG), the primer used for 5′-RACE cDNA synthesis (CD), and the PCR primer (PR) are underlined as described in Methods. The deduced amino acid (aa) sequence contains an 18-aa signal peptide (–20 to –3), a 2-aa proenzyme (–2 to –1), and the mature enzyme (aa 1 to 227). The catalytic triad of amino acid (His45, Asp89, and Ser182), conserved Cys (C) residues, and a conserved sequence for serine proteases at the NH2-terminal (IIGGVE) are in boldface. The sequence is available under GenBank accession number AF063851. (b) Comparison of amino acid sequence of RVCH with other chymases. The deduced amino acid sequence of RVCH is aligned with those of RMCP I, RMCP II, and human chymase (HC). The residues identical to those of RVCH are indicated with hyphens. The conserved catalytic triad, Cys residues, and the conserved sequence for serine proteases (IIGGVE) are in boldface.
Southern blot analysis.
Genomic DNA was extracted from Sprague-Dawley rat hearts using a salt-chloroform method (39). Eight micrograms of DNA was digested with BamHI and EcoRI. The digested DNA was separated on a 0.7% agarose gel, transferred to a Hybond-N+ membrane, and hybridized with the 5′-RACE cDNA fragment or RMCP II cDNA. Hybridization was performed using QuickHyb (Stratagene, La Jolla, California, USA,) according to manufacturer’s instructions.
RVCH expression by RT-PCR.
Rat mast cell lines RBL-1 and RBL-2H3, a rat B-cell line (GP21:56), and a rat fetal fibroblast cell line (Rat2) were obtained from American Type Culture Collection (Rockville, Maryland, USA). Aorta and pulmonary artery SMCs and tissues were taken from 8-week-old Sprague Dawley, SHR, and Wistar-Kyoto (WKY) rats (Charles River Laboratories). Total RNA was extracted from cells and tissues using Trizol as described above. Reverse transcription and PCR conditions were also similar to those described above, but the primers were designed for specific amplification of RVCH cDNA. The forward primer is 5′-GAGGCCTGTAAAATCTATAGAC-3′ and the reverse primer is 5′TGTGTATCTTTGAGAGCCTCAA-3′. The PCR conditions are the same as described above, except that the annealing temperature was set to 58°C, which was optimized to amplify RVCH cDNA specifically. GAPDH cDNA was amplified in a separate PCR reaction as a control for cDNA quality.
Construction of expression vectors and epitope tagging of RVCH.
To maximize translational efficiency, a Kozak consensus sequence (40) GCCACC was added to the RVCH cDNA immediately adjacent to the translation initiation codon using PCR with forward primer 5′-GACTTCAGACTAGTGCCACCATGCAGGCCCTACTATTCCTGATG-3′ and reverse primer 5′-GACTTCAGGCGGCCGCTCAGCTACTTTCCCTTAAGACTGT-3′. The PCR product was subcloned into an expression vector pcDNA3.1(+) (Invitrogen Corp.). The construct obtained was sequenced to verify the integrity of the reading frame and the fidelity of the sequence. To localize RVCH and distinguish recombinant RVCH from endogenous RVCH, HA tag (YPYDVPDYA), a peptide from human influenza hemagglutinin protein, was added to the COOH-terminus of mature RVCH. The two PCR primers used to add HA were as follows: forward primer 5′-GCGGGATCCACTAGTGC-CACCAT-3′; reverse primer 5′-CGGTCTAGATCAAGCGTAGTCTGGGACGTCGTATGGGTAGCTACTTTCCCTTAAGACTGT-3′. The HA sequence is underlined. The resultant PCR product was cloned into pcDNA3.1(+) vector at BamHI/XbaI sites. Sequence analysis was performed to confirm that RVCH with HA tag (RVCH-HA) was in the correct reading frame and no sequence errors had occurred.
In situ hybridization.
To investigate whether RVCH expression would be induced under conditions where there was evidence of active remodeling of the arterial wall associated with proliferation of SMCs, we carried out in situ hybridization using hypertrophied pulmonary artery tissues harvested from rats in which pulmonary hypertension had been induced by an injection of the toxin monocrotaline 21 days earlier (41). Briefly, RVCH was subcloned into pGEM-T vector in a reverse orientation as confirmed by sequencing. Sense and antisense digoxigenin-labeled mRNA probes were synthesized using the DIG RNA labeling Kit (Sp6/T7; Roche Molecular Biochemicals, Laval, Quebec, Canada). Paraffin-embedded sections of pulmonary arteries were hybridized at 65°C for 16 hours in hybridization buffer containing 2 ng/μl digoxigenin-labeled probe, 50% formamide, 1 mg/ml rRNA, and 1× Denhardt’s solution. After washing, sections were blocked at room temperature for 1.5 hours in blocking buffer (pH 7.5; 100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20). Sections were incubated overnight at room temperature with sheep anti-digoxigenin Ab conjugated with alkaline-phosphatase diluted at 1:2,500 in blocking buffer, and then stained using the substrate for alkaline phosphatase.
Northern blot analysis.
Northern blot analysis was performed on samples positive for RVCH expression by RT-PCR, i.e., rat pulmonary artery and aortic SMCs and A10 cells. Twenty-five micrograms of total RNA were separated on a 1% agarose gel and transferred to a Hybond-N+ membrane. The membrane was hybridized with the full-length RVCH cDNA using QuickHyb. After autoradiography, the membrane was then stripped and rehybridized with a rat GAPDH cDNA probe as an internal control.
Localization of RCVH.
Two Ab’s against peptide sequences from RVCH were generated. The peptide sequences were chosen based on specificity, hydrophilicity, antigenic index, and surface probability predicted by MacVector software (Oxford Molecular Group, Oxford, United Kingdom) (42). Peptide A (LSTLQTAYT) and peptide B (GRTGVKDPTSDT) correspond to amino acids 170–179 and 129–140 of mature RVCH, respectively. The Ab against peptide A was prepared in our laboratory, and the Ab against peptide B was prepared by Zymed Laboratories Inc. (South San Francisco, California, USA). Both peptide A and B were coupled to a large carrier protein called keyhole limpet hemocyanin before immunizing the rabbit. The peptide Ab’s were purified using a peptide-affinity column. As assessed by ELISA and dot blot, the rabbits produced a high titer of Ab that specifically recognized the immunizing peptide.
RVCH-HA– and pcDNA-transfected A10 cells, as well as mast cells (RBL-1 and RBL- 2H3, as described above), were fixed using 4% paraformaldehyde followed by a 15-minute permeabilization in PBS containing 0.1% Triton X-100. After blocking with normal serum, cells were incubated with peptide Ab’s (A or B) against RVCH or with a high-affinity rat mAb against the HA tag (1:200). The cells were then incubated with biotinylated anti-rabbit or anti-rat secondary Ab and then with FITC-labeled streptavidin. Normal rabbit or rat IgG was used as a control. Aorta and mesenteric artery from 8-week-old WKY and SHR rats were fixed in neutralized 10% formalin, embedded in paraffin, and used for immunohistochemical staining of chymase. To avoid autofluorescence caused by the abundant elastin in the aorta, sections were stained by an immunoperoxidase method using the Vectastain ABC kit (Vector Laboratories, Burlingame, California, USA), according to the manufacturer’s instructions.
Western blot analysis.
SMCs were obtained from aorta and pulmonary artery of 8-week-old male WKY and SHR rats by explant method. Proteins (10 μg) were resolved by precast 8–16% PAGE (Novex, San Diego, California, USA) and transferred to a PVDF membrane. After blocking, membranes were incubated with RVCH peptide A or B Ab (1:1000). The membranes were washed and incubated with anti-rabbit IgG secondary Ab conjugated with peroxidase (1:10,000). The target protein was detected and visualized by enhanced chemiluminescence according to manufacturer’s instructions.
Transfection procedure.
A10 cells were transfected by Lipofectamine Plus (Life Technologies Inc.) following manufacturer’s instructions. A pcDNA3.1(+) vector, without insert, was used to transfect A10 cells as a control. The recombinant clones were selected in medium containing 400 μg/ml of neomycin (G418) for 3 weeks. The cells were harvested and cell extracts were prepared as described above. It has been shown previously in COS-1 cells that recombinant human heart chymase requires activation by a dipeptidylpeptidase-I (DPPI) (43). To test whether the recombinant rat vascular prochymase was properly processed, the same amount of cell extract was preincubated with 0.1 unit porcine kidney DPPI in 20 mM phosphate buffer, pH 6, containing 0.3 M KCl in a total volume of 20 μl at 37°C for 30 minutes. The cell extract-DPPI reaction mixture was then subjected to a chymase-activity assay using the fluorescent synthetic substrate.
Immunoprecipitation of RVCH.
Recombinant RVCH was immunoprecipitated in HA-tagged RVCH-transfected A10 cells using a kit from Roche Molecular Biochemicals according to the manufacturer’s instruction. High-affinity anti-HA mAb (2 μg/ml) was incubated with cell lysates from pcDNA- and RVCH-HA–transfected A10 cells. Immune complex was collected by centrifugation after incubation with protein G agarose. Immunoprecipitation of HA-tagged RVCH was confirmed by Western blot analysis. Immunoprecipitated samples were resuspended in chymase-activity assay buffer. The ability of recombinant RVCH to cleave Ang I was examined by the same HPLC method used with the A10 cell lysate described below.
Chymase-activity assay.
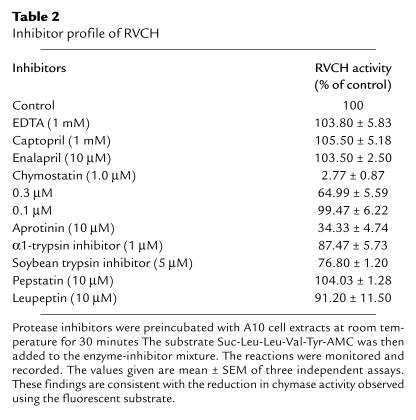
To provide evidence supporting a functionally active RVCH, A10, and aortic and pulmonary artery SMCs from 8-week-old Sprague-Dawley, WKY, and SHR rats (Charles River Laboratories) were used. Samples were prepared according to a paper published previously (44). The vesicles were extracted from SMCs and resuspended in 20 mM Tris-HCl buffer, pH 8.0. A synthetic fluorescent substrate, Suc-Leu-Leu-Val-Tyr-AMC (40), was used to test chymase activity. Chymase activity was defined as fluorescent units per minute per milligram of protein. To further confirm chymase activity, the cell extracts were preincubated at room temperature for 30 minutes with a variety of protease inhibitors, chymostatin, EDTA, captopril, enalapril, aprotinin, soybean trypsin inhibitor, a1-trypsin inhibitor, pepstatin, and leupeptin, in concentrations described previously (20).
Ang II–forming activity related to RVCH was assayed using the protocol described by Urata (20). Briefly, 0.8 fluorescent units per minute enzyme determined by the synthetic substrate were mixed with the reaction buffer. The enzyme-buffer mixture was preincubated with the inhibitors described above or the same volume of inhibitor vehicle, then 10 nM Ang I or Ang II were added. The reactions were incubated at 37°C for 60 minutes and stopped by the addition of 300 μl ice-cold 100% ethanol. The digested peptides were recovered by vacuum drying and analyzed using a C18 reverse-phase HPLC column (Pharmacia, Baie d’Urfe, Quebec, Canada). Peptides were separated using a linear gradient of 12–36% acetonitrile in 0.05% trifluoroacetic acid for 30 minutes at a flow rate of 0.75 ml/min. Elution was monitored as the absorbance at 216 nm. Separated peaks were collected manually and lyophilized. Molecular weight was determined by a mass spectrophotometer.
Calculation of Km and Kcat of purified RVCH.
RVCH was purified from RVCH-transfected A10 SMCs according to methods described previously (27). Briefly, SMC extracts, as described in the chymase-activity assay, were applied to a heparin affinity column (5 ml Hitrap heparin; Amersham Pharmacia Biotech, Piscataway, New Jersey, USA), which was pre-equilibrated with 20 mM Tris-HCl buffer, pH 8.5, containing 0.1 M KCl. The column was eluted with a linear gradient (0–100% for 60 minutes) of 20 mM Tris-HCl buffer, pH 8.0, containing 1.8 M KCl and 0.1% Triton X-100. The flow rate was 0.5 ml/min. These steps were repeated six times, and the active fractions with Ang II–forming activity were pooled. The active fractions were applied to a gel filtration column (1.6 × 60 cm Hiprep Sephacryl S-100 HR; Amersham Pharmacia Biotech), which was equilibrated with 20 mM Tris-HCl, pH 8.5, containing 0.5 M KCl and 0.1% Triton X-100, and then eluted with the same buffer. To determine Km and Kcat values, purified RVCH was incubated with 11 different concentrations (10–600 μM) of Ang I at 37°C for 20 minutes (20). Ang II was separated by HPLC, and its concentration was determined using a radioimmunoassay kit (Peninsula Laboratories Inc., Belmont, California, USA). Km, Vmax and Kcat were calculated using a Lineweaver-Burk plot.
Conversion of Ang I to Ang II in intact SMCs by chymase.
In vivo conversion of Ang I to Ang II was detected in RVCH-transfected A10 cells and SMCs from WKY and SHR rats. RVCH-transfected and vector-transfected A10 cells, as well as SMCs from SHR and WKY rats, were incubated with Ang I (1 pM) for 1 hour. In some experiments, cells were preincubated with chymase inhibitor chymostatin for 15 minutes before the addition of Ang I. SMCs were harvested, and cell homogenates were centrifuged at 10,000 g for 15 minutes. The resulting supernatant was used for detection of Ang II. Ang II was separated by HPLC, as described above, and Ang II concentration was measured by radioimmunoassay kit (Peninsula Laboratories Inc.).
Statistical analysis.
All data are expressed as mean ± SEM. The difference between two groups was calculated using Student’s t test. ANOVA analysis was used when three groups were compared followed by a Tukey’s test of multiple comparisons. A P value of less than 0.05 was considered statistically significant.
Results
Cloning and sequencing of RVCH cDNA.
We generated cDNA sequences that might encode novel proteolytic enzymes in rat pulmonary artery SMCs. The 3′-RACE PCR products of the expected size range of serine proteases, i.e., between 300 and 500 bp, were isolated and subcloned, and 90 colonies were sequenced and compared with the GenBank database. Among these sequences, clone 3–19 contained an insert of 311 bp with 84% homology to the 3′ end of RMCP II cDNA. The sequence contained an open reading frame encoding 44 amino acid (aa), a stop codon, and a 177-bp 3′ untranslated region containing a putative polyadenylation signal sequence AAUAAA. From this sequence, two primers were designed and used to clone the 5′ end of the cDNA, one for cDNA synthesis and one for PCR amplification as described in Methods. Use of 5′-RACE resulted in a 777-bp sequence with 135 bp overlapping with clone 3–19. The full-length cDNA (Figure 1a) was obtained by PCR with primers at the ends of 3′-RACE and 5′-RACE fragments.
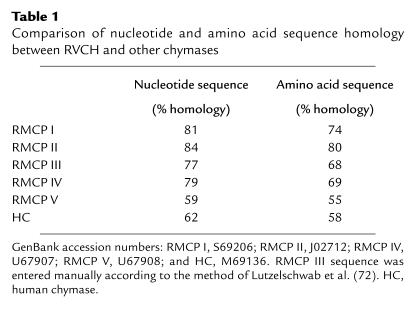
The cDNA encompasses 953 bp with a 33-bp 5′ untranslated region, 740-bp coding region, and a 177-bp 3′ untranslated region described above, and the sequence was confirmed in three different colonies and in RT-PCR products from rat pulmonary artery SMCs, as well as A10 cells. The 247 deduced amino acid sequence is composed of an 18-aa signal peptide, a 2-aa proenzyme, and a 227-aa mature enzyme containing the conserved catalytic triad, His45, Asp89, and Ser182, and six conserved cysteines. A GenBank search reveals that RVCH is a novel gene with high homology to other members of the chymase family, e.g., 81 and 84% nucleotide and 74 and 80% amino acid homology to RMCP I and RMCP II, respectively (Figure 1b; Table 1).
Table 1.
Comparison of nucleotide and amino acid sequence homology between RVCH and other chymases
Southern blot analysis of RVCH.
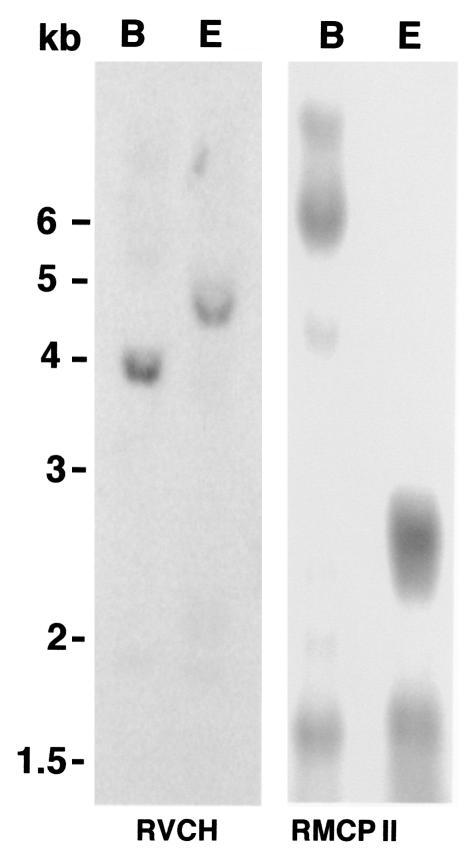
To confirm that RVCH is derived from a gene distinct from that of other chymases, Southern blot analysis was carried out. Rat genomic DNA was digested with BamHI and EcoRI and probed with the 5′-RACE fragment of RVCH and RMCP II cDNA. When stringency was increased by raising the washing temperature to 68°C, only a 3.8-kb BamHI and a 4.5 kb EcoRI band were detectable (Figure 2). The Southern blot pattern for RVCH is different from that detected using RMCP II cDNA, i.e., a 6-kb BamHI restriction fragment and multiple EcoRI fragments range from 1.5 to 2.2 kb, as reported previously (45). Thus, analysis of genomic DNA indicates that RVCH is encoded by a separate gene but there is cDNA homology to other members of the chymase family.
Figure 2.
Southern blot analysis of the RVCH gene. Rat genomic DNA was digested with BamHI (B) and EcoRI (E). The blot was hybridized with the 5′-RACE fragment of RVCH and RMCP II cDNA. The membrane was washed with 2× SSC/0.1% at room temperature, followed by SDS 0.1× SSC/0.1% SDS at 68°C. The pattern of bands detected by the 5′-RACE fragment of RVCH was different from that detected with RMCP II cDNA.
Expression of RVCH mRNA.
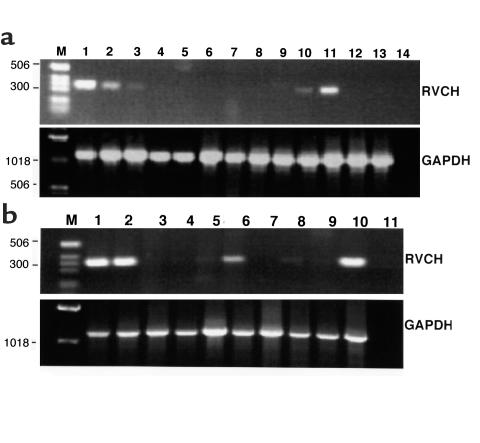
RT-PCR was performed using RNA from different tissues and cell lines to investigate the pattern of RVCH expression. RVCH mRNA was detected in pulmonary artery and aortic SMCs from the Sprague-Dawley rats, A10 cells, mast cell lines (RBL-1 and RBL-2H3), but not in pulmonary artery, aorta, or other tissues screened, such as heart, lung, kidney, liver, or skeletal muscle, and not in a rat B-cell line (GP21:56) or a rat fetal fibroblast cell line (Rat 2) (Figure 3a). We further investigated whether primary cultured SMCs and tissues derived from SHR expressed RVCH. When RT-PCR was performed with RNA from different SHR tissues, RVCH transcripts were found in cultured pulmonary artery and aortic SMCs and lung tissue but not in aorta, pulmonary artery, or other tissues, which were also negative in Sprague-Dawley rats (Figure 3b). We showed, however, by in situ hybridization, upregulation of RVCH mRNA associated with evidence of SMC proliferation in hypertrophied pulmonary arteries harvested from rats 21 days after injection of the toxin monocrotaline (Figure 4f), but not in control vessels from rats injected with saline (Figure 4c).
Figure 3.
Expression of the RVCH in different tissues and cells. (a) RT-PCR was performed on mRNA from Sprague-Dawley pulmonary artery (PA) SMCs (lane 1), aorta (AO) SMCs (lane 2), A10 cells (lane 3), PA (lane 4), AO (lane 5), heart (lane 6), kidney (lane 7), lung (lane 8), muscle (lane 9), rat mast cell line RBL-1 (lane 10), rat mast cell line RBL-2H3 (lane 11), rat B-cell line GP21:56 (lane 12), rat fetus fibroblast cell line Rat 2 (lane 13), and negative control (lane 14). (b) SHR cells and tissues visualized were: PA SMCs (lane 1), AO SMCs (lane 2), PA (lane 3), AO (lane 4), heart (lane 5), lung (lane 6), kidney (lane 7), liver (lane 8), muscle (lane 9), and Sprague-Dawley PA SMCs (lane 10) as a positive control and negative control (lane 11). GAPDH cDNA was amplified as a control for cDNA quality. The base-pair sizes of commercially prepared markers (lane M) are indicated on the left.
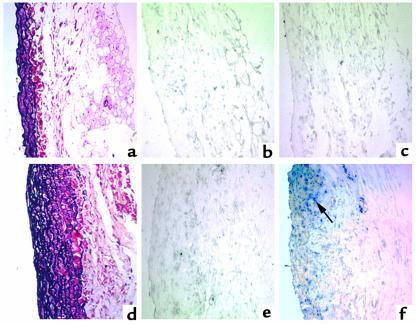
Figure 4.
In situ hybridization analysis of RVCH mRNA expression in pulmonary hypertension. Representative photomicrographs of in situ hybridization with digoxigenin-labeled RVCH antisense (c and f) and sense (b and e) riboprobes in PA from rats 21 days after monocrotaline (d, e, and f) and saline injection (a, b and c). RVCH mRNA expression appears as dark blue staining (arrow) and is only detectable in monocrotaline-treated rats. Movat pentachrome staining shows PA hypertrophy in monocrotaline-treated rats (d) compared with saline-treated rats (a).
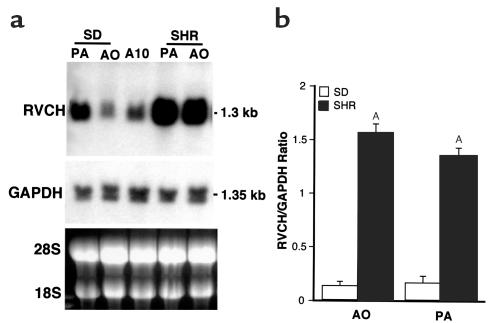
To determine the levels of expression of RVCH in vascular SMCs, Northern blot analyses were carried out. The full-length RVCH cDNA when used as a probe, hybridized to a single band of 1.3 kb in RNA from pulmonary artery and aortic SMCs of Sprague-Dawley and SHR rats, as well as from A10 cells (Figure 5a). The Northern blot results were consistent in RNA samples from three independent primary cultures, compared by densitometry and normalized to GAPDH. An 11-fold and more than 8-fold increase in RVCH mRNA levels were detected, respectively, in aorta and pulmonary artery SMCs of SHR rats when compared with Sprague-Dawley rats (Figure 5b). A similar result was found when comparing RNA from SHR with genetically related WKY rats (data not shown). To confirm sequence identity of RVCH in SHR rats, we amplified by PCR and subcloned the full-length RVCH cDNA from pulmonary artery SMCs of SHR rats. No mutation, that might account for the increased expression of the mRNA was found by sequencing of the complete cDNA from three colonies and from the RT-PCR products.
Figure 5.
Northern blot analysis of RVCH mRNA levels. Total RNA from Sprague-Dawley PA SMCs, AO SMCs, A10 cell line, SHR PA SMCs, and AO SMCs was used for Northern blot analysis of RVCH mRNA levels. (a) A representative blot showed a single 1.3-kb mRNA band in all samples detected using the full-length RVCH cDNA (953 nucleotide) probe. The membrane was then stripped and rehybridized with a GAPDH probe as a control. (b) Densitometric quantification of Northern blots revealed a significant increase in the RVCH/GAPDH ratio in SHR versus Sprague-Dawley in both AO and PA. The data are depicted as mean ± SEM of three independent experiments. AP < 0.05 versus Sprague-Dawley.
Expression of RVCH protein.
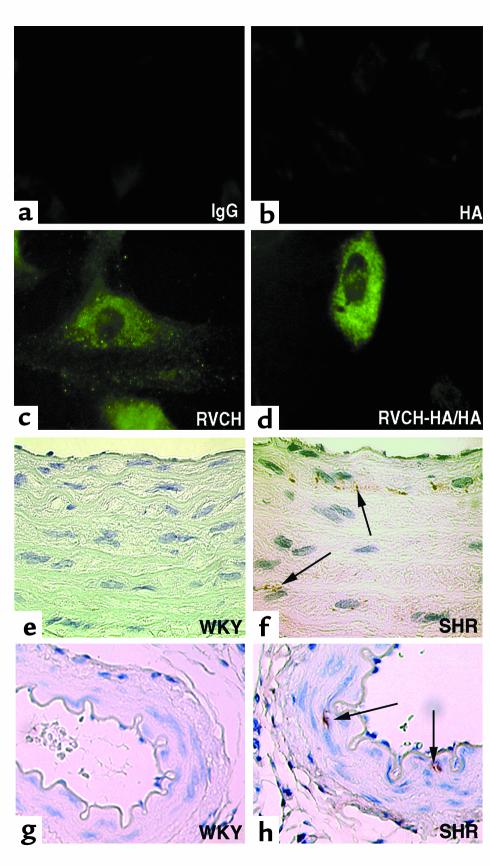
Mast cell chymase described previously has been localized intracellularly to mast cell granules. Immunofluorescent staining using a chymase peptide Ab (B) revealed a punctate pattern of immunostaining in the cytosol of control vector–transfected cells, which was similar to that observed using a high-affinity mAb against HA in RVCH-HA tag–transfected A10 cells (Figure 6, c and d). The punctate staining disappeared when the peptide Ab was preincubated with 20 times more of the corresponding synthetic peptide. The similar distribution of recombinant-tagged RVCH and endogenous RVCH immunostaining in the cytosol is consistent with both reflecting the same protein. Furthermore, no staining was found when the same peptide Ab was applied to cultured mast cell lines RBL-1 and RBL-2H3 (data not shown). Chymase was also detected by immunoperoxidase staining in SMCs of the aorta from SHR but not WKY rats (Figure 6, f and h). The staining is specific for RVCH; it was completely blocked following preincubation of the peptide Ab with synthetic peptide.
Figure 6.
(a–d) Localization of endogenous RVCH and recombinant RVCH-HA tag in transfected A10 cells. Staining with the peptide Ab (c) or an anti-HA tag Ab (d) revealed a similar punctate immunofluorescent staining in cytosol of representative A10 cells. As expected, the intensity of immunostaining was higher in the RVCH-HA–transfected (RVCH-HA/HA) than in the vector-transfected cell, stained with the anti-HA Ab and Ab against peptide A, respectively. A similar result was obtained with Ab against peptide B. Negative controls are shown using rabbit IgG (a) and an Ab to HA tag (b) in nontransfected cells. ×100. (e–h) Immunohistochemical staining of RVCH in AO and mesentery artery from WKY and SHR rats. Immunoperoxidase staining of AO (f) and mesentery artery (h) sections reveals immunodetection of RVCH (arrows) in medial SMCs in 8-week-old SHR rats, but no staining was found in an AO (e) or mesentery artery (g) from an age-matched WKY rat. ×100. These results were obtained using the Ab against peptide B, described in the text. Representative of n = 3 per group.
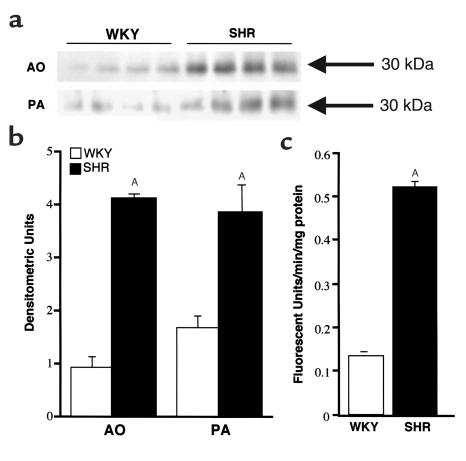
Western blot analysis using the peptide Ab specific to RVCH detected a 30-kDa band in SMC extracts (Figure 7a), consistent with the size of RVCH. Densitometric analysis of this band revealed that RVCH was significantly increased in SMCs from the aorta and pulmonary artery of SHR compared with WKY rats (4.5- and 2.3-fold, respectively) (Figure 7b).
Figure 7.
Western blot analysis shows increased RVCH in SMC from SHR versus WKY rats. Total protein (10 mg) was resolved using an 8–16% gradient Tris-glycine gel. A peptide Ab that is specific to RVCH was used as the primary Ab. A similar result was obtained with the Ab against peptide A or B. (a) A representative Western blot showed a specific band for 30-kDa RVCH. (b) Densitometric evaluation of RVCH in SMCs reveals an increase in SHR versus WKY rats. (c) Chymase activity was increased in aortic SMCs from SHR versus WKY rats. The data are depicted as the mean ± SEM of four experiments. AP < 0.05 versus WKY-transfected, but not pcDNA-transfected (vector alone), A10 cells.
Demonstration of chymase activity in SMCs.
The increase in chymase protein concentration in SMC extracts from SHR versus WKY rats was reflected in a similar elevation in chymase activity (Figure 7c), detected as described in the Methods, using a selective synthetic fluorescent substrate (Suc-Leu-Leu-Val-Tyr-AMC) and recorded as an increasing amount of free AMC over time. We also detected chymase activity in A10 cell extracts, but not in conditioned media, indicating that it was not secreted under normal culture conditions. To confirm that this reaction was specific for chymase activity, different protease inhibitors were preincubated with the A10 cell extracts. The enzyme activity was almost completely inhibited by chymostatin, a chymase inhibitor, partially inhibited by other serine protease inhibitors such as aprotinin, α1-trypsin inhibitor, and soybean trypsin inhibitor, but not by the general protease inhibitor pepstatin, the metalloproteinase inhibitor, EDTA, the aspartic protease inhibitor, leupeptin, or by the ACE inhibitors, captopril and enalapril (Table 2). As well, chymase activity was observed in primary SMCs cultured from rat, porcine, and human aorta and pulmonary artery (data not shown).
Table 2.
Inhibitor profile of RVCH
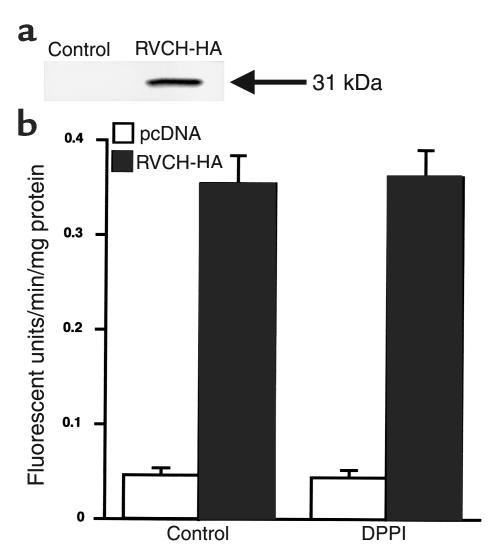
To confirm that the RVCH cDNA specifically encodes a functionally active chymase, we expressed recombinant RVCH in A10 cells and tested chymase activity using Suc-Leu-Leu-Val-Tyr-AMC. The expression (Figure 8a) and successful immunoprecipitation of HA-tagged RVCH was confirmed by Western blot analysis. A10 cells stably transfected with the RVCH cDNA construct showed 8.5-fold increase in chymase activity compared with the control vector (Figure 8). Addition of DPPI did not cause a further increase in chymase activity in both RVCH and control vector–transfected cells, indicating that A10 cells have the ability to process the recombinant pro-chymase to the active enzyme.
Figure 8.
Expression of recombinant RVCH in A10 cells. (a) The expression of recombinant RVCH was detected by Western blot analysis in cell lysates of HA-tagged RVCH-transfected, but not pcDNA-transfected, A10 cells, using a high-affinity mAb against HA (RVCH + HA = 31 kDa). (b) Chymase activity increased 8.5-fold in A10 cells stably transfected with RVCH cDNA construct compared with those transfected with the control vector pcDNA. Chymase activity does not increase after addition of 0.1 units of DPPI in A10 cells transfected with the RVCH cDNA construct or in control vector–transfected cells. The data are depicted as mean ± SEM of three independent experiments. P < 0.05, RVCH versus pcDNA.
Ang II formation.
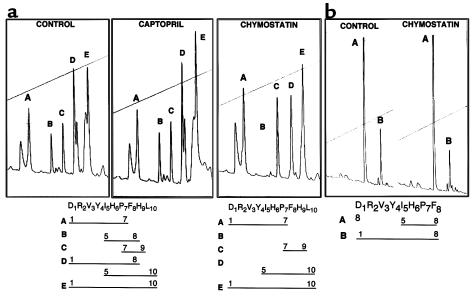
We next determined whether SMC chymase activity could result in Ang II conversion from Ang I. Ang I was used as a substrate, and after incubation with A10 cell extracts, HPLC analysis was used to detect reaction products. Five major peaks were observed (Figure 9a), and the Ang I-derived peptides were identified using mass spectrophotometry. Peak A contained peptide Asp1-Pro7; peak B, peptide Ile5-Phe8; peak C, Pro7-His9; peak D, Ang II (Asp1-Phe8) and Ile5-Leu10; and peak E, undigested Ang I (Asp1-Leu10). To distinguish the peptides that were specifically generated by chymase activity, A10 cell extracts were preincubated with different protease inhibitors. When chymostatin was added to the cell extracts, peak B (fragment Ile5-Phe8) and the Ang II component in peak D were completely eliminated. Ang II formation detected by HPLC was inhibited by chymostatin, but not by captopril (Figure 9a) or enalapril (not shown), suggesting that the formation of Ang II from Ang I in these cultured A10 cells resulted from chymase rather than ACE activity. There was some inhibition of Ang II peptide formation by aprotinin, α1-trypsin inhibitor, and soybean trypsin inhibitor, judged by analysis of HPLC peaks, although not to the extent observed with chymostatin, and there was no inhibition observed with EDTA, leupeptin, or pepstatin (data not shown). The fragment Ile5-Phe8 shares a common cleavage site at the Phe8-His9 bond with Ang II. It is this cleavage site that is attributable to chymase activity.
Figure 9.
(a) Conversion of Ang II from Ang I by A10 cell extracts. Ang I was incubated with A10 cell extracts and analyzed using HPLC. The separated peaks were collected for molecular-weight analysis by mass spectrophotometer. The fragments contained in different peaks are illustrated below the figures. Chymostatin eliminated peak B (Ile5-His6-Pro7-Phe8) and the Ang II component of peak D (Asp1 to Phe8). Captopril could not inhibit the generation of Ang II, suggesting that the cleavage at the Phe8-His9 bond is the result of a chymase rather than angiotensin-converting enzyme. (b) Cleavage of the Tyr4-Ile5 bond in Ang II cannot be inhibited by chymostatin. Ang II was incubated with A10 cell extracts and analyzed by HPLC. Peak A contains Ile5-His6-Pro7-Phe8. Peak B contains Ang II (Asp1 to Phe8). Addition of chymostatin cannot inhibit cleavage of the Tyr4-Ile5 bond.
We further tested directly Ang II cleavage by chymase by adding Ang II to A10 cell extracts and analyzed the reaction products using HPLC (Figure 9b). Ang II was cleaved at the Tyr4-Ile5 bond, but this cleavage could not be inhibited by 10 μM or 100 μM chymostatin synthetic substrate. The Ile5-Leu10 peptide in peak D was protected from cleavage by chymostatin. This suggests that Tyr4-Ile5 cleavage may be mediated by other proteinases in the cell lysate.
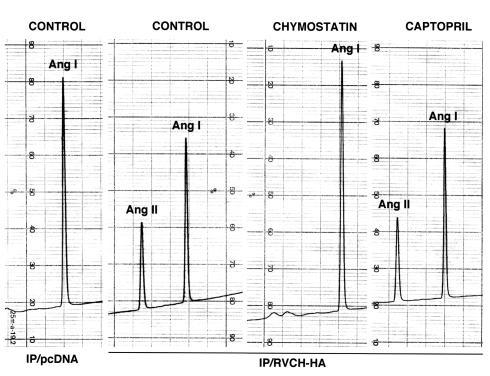
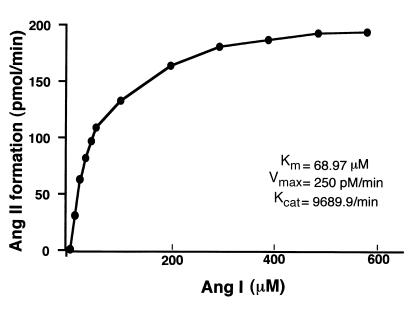
These results were further confirmed by the conversion of Ang I to Ang II when Ang I was incubated with immunoprecipitated HA-tagged RVCH (Figure 10). We did not observe peaks B (Ile5-Phe8) and D (Ile5-Leu10) due to cleavage of Tyr4-Ile5, further supporting lack of cleavage by RVCH. Our results suggested that this novel vascular chymase has α-chymase function. This was further supported by a similar Km (68.97 μM), Vmax (250 pM/min), and Kcat (9689.9/min) values (Figure 11) compared with α-chymase (human chymase) (20).
Figure 10.
Conversion of Ang I to Ang II by immunoprecipitated recombinant RVCH. Cell lysates from pcDNA and HA-tagged RVCH-transfected A10 cells were immunoprecipitated with high-affinity mAb against HA. Ang I was incubated with immunoprecipitated RVCH and analyzed using HPLC. The separated peaks were collected for molecular-weight analysis by mass spectrophotometry. Conversion of Ang I to Ang II was confirmed only when Ang I was incubated with immunoprecipitated protein from HA-tagged RVCH. Chymostatin eliminated the Ang II peak. Captopril could not inhibit the generation of Ang II, suggesting that the cleavage at the Phe8-His9 bond is the result of a chymase rather than angiotensin-converting enzyme.
Figure 11.
The kinetics of Ang II formation from Ang I by purified vascular chymase. Purified chymase was incubated with 10–600 μM Ang I at 37°C for 20 minutes. Ang II generated was analyzed using HPLC, followed by radioimmunoassay. Values shown in this figure represent the mean of four independent experiments. Km, Vmax, and Kcat were calculated using the Lineweaver-Burk plot.
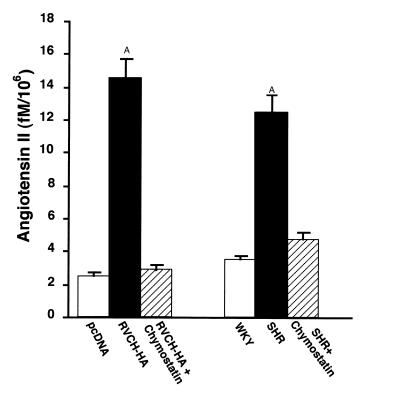
The chymase-dependent conversion of Ang I to Ang II was confirmed in RVCH-HA–transfected A10 cells and SMCs from SHR rats. It was of interest that all the increase in Ang II production in the RVCH-transfected cells, when compared with control, as well as in the SHR compared with WKY SMCs, was chymostatin inhibitable (Figure 12), indicating that chymase is responsible for Ang I to Ang II conversion in SMCs.
Figure 12.
Conversion of Ang I to Ang II by intact SMCs. RVCH-HA–transfected and vector-transfected A10 cells, as well as SMCs from SHR and WKY rats, were incubated with Ang I (1 pM) for 1 hour. In some experiments, SMCs were preincubated with the chymase inhibitor, chymostatin, for 15 minutes before the addition of Ang I. SMCs were harvested, and Ang II was separated using HPLC subsequent to quantification by radioimmunoassay. Values given are mean ± SEM of four independent experiments. AP < 0.05 versus all other groups.
Discussion
We cloned a full-length cDNA encoding a novel chymase from cultured rat vascular SMCs. This chymase shows high homology in cDNA and amino acid sequence to RMCP I and RMCP II produced by mast cells. The cDNA cloned was transcribed from a separate gene; a 1.3-kb mRNA was observed in RNA from primary cultured pulmonary artery and aorta SMCs and in the A10 cell line, and there was a marked increase in expression in cells from SHR versus normotensive rats. In situ hybridization also revealed increased RVCH expression in arteries from rats with pulmonary hypertension. Both endogenous and recombinant RVCH protein show a similar immunolocalization pattern in the cytosol, and an increase in RVCH detected in the aorta of SHR versus normotensive rats correlates with an elevation in protein levels assessed by Western immunoblotting and in chymase activity assessed using an immunofluorescent substrate. Transfecting A10 cells with the RVCH cDNA construct further documented that the translated product is a functionally active chymase. Conversion of Ang I to Ang II was documented by HPLC separation and analysis of peptides by mass spectrophotometry, using cell lysates, purified recombinant RVCH, and intact SMCs. Moreover, in SMCs from SHR versus WKY rats, there was a marked increase in Ang II conversion from Ang I, which was inhibited by chymostatin. Our results therefore provide the first evidence, to our knowledge, that links a specific cDNA sequence encoding RVCH to a functionally active chymase in vascular SMCs. In SHR versus normotensive rats, the increase in RVCH mRNA and protein in smooth muscle cells correlates with chymase activity and angiotensin conversion, suggesting that RVCH may be related to hypertension.
Using RT-PCR, we showed that RVCH was expressed in vascular SMCs as well as in mast cell lines (RBL-1, RBL-2H3), but not in a B-cell line (GP21:56) and a fibroblast cell line (rat2), excluding the possibility that the gene is constitutively expressed in all proliferating cells. The contamination of primary SMCs by mast cells can be excluded since these cultures have been shown to uniformly express smooth muscle α-actin and since RVCH mRNA was also detected in the aortic SMC line (A10). In Sprague-Dawley rat tissues, e.g., pulmonary artery, aorta, and heart, the level of RVCH transcripts was below detection by RT-PCR, suggesting that the mRNA may be transcribed only in actively proliferating cultured SMCs. This is consistent with the increase in RVCH mRNA expression by in situ hybridization of hypertrophied but not normal pulmonary arteries. We speculate that RVCH mRNA might also be induced during a certain window in development or in response to arterial injury, e.g., in atherosclerosis when SMCs are relatively dedifferentiated, proliferating, and migrating. It is possible that in mature vascular and other tissues the enzyme is stored in granules like other chymases in mast cells. Another example is neutrophil elastase mRNA, which can be detected in immature promyelocytes but not in mature neutrophils that store the active enzyme in granules (46). Mast cell proteases are bound to proteoglycans in secretory granules and released only when cells are stimulated (47). This may explain why, in some reports, chymostatin-sensitive Ang II–forming activity was undetectable in rat arteries under physiological condition (23).
Chymases are potent proteases with numerous target substrates. Known chymase functions include conversion of big-endothelin-1 to endothelin-1 (48), inactivation of bradykinin and kallidin (49), degradation of substance P and vasoactive intestinal peptide (50), and formation of Ang II from Ang I. Chymases may also play an important role in cardiovascular tissue remodeling through their ability to activate progelatinase B (MMP-9) (51, 52), to directly degrade matrix proteins such as fibronectins and type IV collagen (53, 54), and to initiate collagen fibril formation by cleaving type I procollagen. Chymases can also stimulate collagen synthesis and SMC proliferation by converting Ang I to Ang II (55). Ang I catalysis may differ in human and rat chymases. Human chymase, specifically, hydrolyses the Phe8-His9 bond of Ang I, while RMCP I degrades the Tyr4-Ile5 bond with approximately 20-fold higher catalytic efficiency than the Phe8-His9 of Ang I (56).
Mammalian chymases can be classified as two subgroups according to their structure and substrate specificity (57). The α-chymases include human chymase, baboon chymase, dog chymase, and mouse chymase-5 and convert Ang I to Ang II, while not degrading Ang II. The remaining known mammalian chymases, e.g., RMCP I, RMCP II, mouse chymase -1, -2, -4, and gerbil chymase-1 are classified as β-chymases. The novel vascular chymase, RVCH, shows 74% and 79% homology in amino acid sequence to RMCP I and RMCP II and 56% homology to human heart chymase, thus, structurally belongs to the β-chymase family. We tested chymase activity in A10 cell extracts using Ang I and II as substrates. The cleavage of the Phe8-His9 bond was completely inhibited by chymostatin, while cleavage of the Tyr4-Ile5 bond was not. This raises the possibility that the cleavage of the Tyr4-Ile5 bond could result from other enzymes in the cell extracts. This result was supported by the observation that purified RVCH only cleaves Ang I at Phe8-His9. Based on the substrate specificity and similar Km and Kcat value to human chymase (α-chymase), this vascular chymase has α-chymase function indicating functional heterogeneity in chymases classified structurally as members of the β subgroup.
We showed through transfection studies that the RVCH cDNA can be translated to a functionally active chymase and processed correctly in A10 cells. Based on the deduced amino acid sequence, RVCH contains an 18-aa signal peptide and a 2-aa proenzyme peptide Glu-Glu. As reported previously, Glu-Glu is a cleavage site for DPPI (58). It appears, like other chymases, to be translated initially as a zymogen and then processed to an active form by endogenous DPPI cleavage of the propeptide (59, 60). Since transfection of COS-1 cells with the human chymase results in an inactive prochymase (43), we selected A10 cells for transfection. A10 cells have chymase activity and therefore should possess mechanisms necessary for processing the prochymase. The addition of DPPI to the cell extracts resulted in only a minimal increase in chymase activity, indicating that most of the recombinant RVCH in A10 cells was processed properly. Our preliminary data reveal that TGF-β1, Il-10, and TNF-α do not stimulate the release of chymase from cultured A10 cells (unpublished data). Thus, the regulation of chymase release in SMCs needs further study.
Since both peptide Ab’s fail to immunoprecipitate RVCH, we cannot assess precisely the contribution of RVCH to the chymase activity detected in the cell extracts. Our results, however, are consistent with a relationship between the cloned RVCH and endogenous SMC chymase based upon the following experimental evidence. First, transfection of A10 cells with cloned RVCH increases chymase activity in cell extracts; second, an endogenous protein that has the same molecular weight as chymase was detected by using an Ab specific to RVCH; third, a similar distribution pattern was revealed by immunostaining using Ab’s against endogenous RVCH and the transfected RVCH with the HA tag.
The SHR is the most widely studied animal model of essential hypertension. Although the exact mechanism of hypertension in SHR rats is not clear, it is generally accepted that increased local production of Ang II from cardiovascular tissues plays a crucial role in the initiation and maintenance of hypertension in this model (61, 62). While the pathophysiological role of RVCH in the development of hypertension in the SHR rat remains to be determined, it could potentially account for the elevation in Ang II.
The RVCH mRNA levels are elevated in cultured aorta SMCs from SHR when compared with those from Sprague-Dawley and WKY rats. This suggests that the abnormality may be related to enhanced transcriptional regulation or stability of chymase mRNA. The increase in RVCH mRNA in cultured vascular cells, but not in vascular tissue from adult rats, is intriguing, and studies addressing the temporal expression of this enzyme in proliferating cells in embryonic and neonatal vessels would be of interest, particularly if a correlation could be made with the onset of hypertension. It is of interest that a similar increase in Ang II production from Ang I was detected in the cells of SHR versus WKY rats, as well as in RVCH versus vector-transfected A10 cells, and could be attributed to chymase activity, i.e., in that it was chymostatin inhibitable. It has been shown in hepatocytes (63, 64) and in cancer cells (65) that chymostatin can enter cells and inhibit intracellular proteolysis, and chymase inhibitors can also inhibit enzymes associated with the cell membrane (66). Recent studies have shown that intracellular Ang II can play an important role in gene regulation and cell communication (67) via an intracellular receptor (68, 69) and that its release may be regulated by mechanical factors (70).
It has been reported that the pulmonary pressure is also elevated in SHR as compared with WKY rats (71). This result is consistent with our finding of increased expression of chymase in pulmonary vascular SMCs from SHR rats. We also found evidence of upregulation of RVCH mRNA in hypertrophied pulmonary arteries from rats 21 days after injection of monocrotaline. Our previous studies had shown active remodeling with evidence of vascular SMC proliferation and accumulation of ECM (41). In addition to RVCH mRNA levels, chymase protein and enzyme activity related to production of Ang II are elevated in SHR vascular cells, and this could be attributed to RVCH and non-RVCH chymases. Taken together, our results suggest only that RVCH might be relevant to the pathophysiology of hypertension in the SHR rats. To further investigate this relationship it will be of interest to selectively inhibit RVCH in SHR rats through their development or to overexpress RVCH in transgenic mice.
Acknowledgments
We are indebted to Kyle Northcote Cowan, Chi-Chung Hui, and Rong Mo for providing sample and technical assistance with in situ hybridization. We wish to thank Lars Hellman from University of Uppsala, Sweden, for providing RMCP II cDNA. We are grateful to Chris Howard for assistance in preparing the plasmid for Southern blot analysis and Joan Jowlabar and Judy Matthews for secretarial assistance. This work is supported by grant T3170 from the Heart and Stroke Foundation of Ontario. M. Rabinovitch is a Research Endowed Chair of the Heart and Stroke Foundation of Ontario and a Distinguished Scientist of the Medical Research Council of Canada. H. Ju and H. Massaeli are Research Fellows of the Medical Research Council of Canada and the Heart and Stroke Foundation of Canada.
Footnotes
Caiying Guo and Haisong Ju contributed equally to this work.
References
- 1.Morrell NW, Atochina EN, Morris KG, Danilov SM, Stenmark KR. Angiotensin converting enzyme expression is increased in small pulmonary arteries of rats with hypoxia-induced pulmonary hypertension. J Clin Invest. 1995;96:1823–1833. doi: 10.1172/JCI118228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrell NW, Morris KG, Stenmark KR. Role of angiotensin-converting enzyme and angiotensin II in development of hypoxic pulmonary hypertension. Am J Physiol. 1995;269:H1186–H1194. doi: 10.1152/ajpheart.1995.269.4.H1186. [DOI] [PubMed] [Google Scholar]
- 3.Danser AH, Schalekamp MA. Is there an internal cardiac renin-angiotensin system? Heart. 1996;76(Suppl. 3):28–32. doi: 10.1136/hrt.76.3_suppl_3.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosendorff C. Vascular hypertrophy in hypertension: role of the renin-angiotensin system. Mt Sinai J Med. 1998;65:108–117. [PubMed] [Google Scholar]
- 5.Curzen NP, Fox KM. Do ACE inhibitors modulate atherosclerosis? Eur Heart J. 1997;18:1530–1535. doi: 10.1093/oxfordjournals.eurheartj.a015132. [DOI] [PubMed] [Google Scholar]
- 6.Brilla CG, Scheer C, Rupp H. Renin-angiotensin system and myocardial collagen matrix: modulation of cardiac fibroblast function by angiotensin II type 1 receptor antagonism. J Hypertens Suppl. 1997;15:S13–S19. [PubMed] [Google Scholar]
- 7.Baker KM, Aceto JF. Angiotensin II stimulation of protein synthesis and cell growth in chick heart cells. Am J Physiol. 1990;259:H610–H618. doi: 10.1152/ajpheart.1990.259.2.H610. [DOI] [PubMed] [Google Scholar]
- 8.Daemen MJ, Lombardi DM, Bosman FT, Schwartz SM. Angiotensin II induces smooth muscle cell proliferation in the normal and injured rat arterial wall. Circ Res. 1991;68:450–456. doi: 10.1161/01.res.68.2.450. [DOI] [PubMed] [Google Scholar]
- 9.Intengan HD, Thibault G, Li JS, Schiffrin EL. Resistance artery mechanics, structure, and extracellular components in spontaneously hypertensive rats: effects of angiotensin receptor antagonism and converting enzyme inhibition. Circulation. 1999;100:2267–2275. doi: 10.1161/01.cir.100.22.2267. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer MA, et al. Effect of captopril on mortality and morbidity in patients with left ventricular dysfunction after myocardial infarction. Results of the survival and ventricular enlargement trial. The SAVE Investigators. N Engl J Med. 1992;327:669–677. doi: 10.1056/NEJM199209033271001. [DOI] [PubMed] [Google Scholar]
- 11.Pfeffer JM, Pfeffer MA, Mirsky I, Braunwald E. Regression of left ventricular hypertrophy and prevention of left ventricular dysfunction by captopril in the spontaneously hypertensive rat. Proc Natl Acad Sci USA. 1982;79:3310–3314. doi: 10.1073/pnas.79.10.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chobanian AV, Haudenschild CC, Nickerson C, Hope S. Trandolapril inhibits atherosclerosis in the Watanabe heritable hyperlipidemic rabbit. Hypertension. 1992;20:473–477. doi: 10.1161/01.hyp.20.4.473. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki M, Sakonjo H, Takai S. Anti-atherosclerotic effects of an angiotensin converting enzyme inhibitor and an angiotensin II antagonist in Cynomolgus monkeys fed a high-cholesterol diet. Br J Pharmacol. 1999;128:523–529. doi: 10.1038/sj.bjp.0702833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okunishi H, Miyazaki M, Toda N. Evidence for a putatively new angiotensin II-generating enzyme in the vascular wall. J Hypertens. 1984;2:277–284. [PubMed] [Google Scholar]
- 15.Gondo M, Maruta H, Arakawa K. Direct formation of angiotensin II without renin or converting enzyme in the ischemic dog heart. Jpn Heart J. 1989;30:219–229. doi: 10.1536/ihj.30.219. [DOI] [PubMed] [Google Scholar]
- 16.Urata H, Healy B, Stewart RW, Bumpus FM, Husain A. Angiotensin II-forming pathways in normal and failing human hearts. Circ Res. 1990;66:883–890. doi: 10.1161/01.res.66.4.883. [DOI] [PubMed] [Google Scholar]
- 17.Shiota N, et al. Activation of angiotensin II-forming chymase in the cardiomyopathic hamster heart. J Hypertens. 1997;15:431–440. doi: 10.1097/00004872-199715040-00014. [DOI] [PubMed] [Google Scholar]
- 18.Hoit BD, et al. Effects of angiotensin II generated by an angiotensin converting enzyme-independent pathway on left ventricular performance in the conscious baboon. J Clin Invest. 1995;95:1519–1527. doi: 10.1172/JCI117824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urata H, et al. Cloning of the gene and cDNA for human heart chymase. J Biol Chem. 1991;266:17173–17179. [PubMed] [Google Scholar]
- 20.Urata H, Kinoshita A, Misono KS, Bumpus FM, Husain A. Identification of a highly specific chymase as the major angiotensin II-forming enzyme in the human heart. J Biol Chem. 1990;265:22348–22357. [PubMed] [Google Scholar]
- 21.Urata H, et al. Cellular localization and regional distribution of an angiotensin II-forming chymase in the heart. J Clin Invest. 1993;91:1269–1281. doi: 10.1172/JCI116325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urata H, Strobel F, Ganten D. Widespread tissue distribution of human chymase. J Hypertens Suppl. 1994;12:S17–S22. [PubMed] [Google Scholar]
- 23.Okunishi H, et al. Marked species-difference in the vascular angiotensin II-forming pathways: humans versus rodents. Jpn J Pharmacol. 1993;62:207–210. doi: 10.1254/jjp.62.207. [DOI] [PubMed] [Google Scholar]
- 24.Shiota N, et al. Activation of two angiotensin-generating systems in the balloon-injured artery. FEBS Lett. 1993;323:239–242. doi: 10.1016/0014-5793(93)81348-4. [DOI] [PubMed] [Google Scholar]
- 25.Mangiapane ML, et al. Vasoconstrictor action of angiotensin I-convertase and the synthetic substrate (Pro11,D-Ala12)-angiotensin I. Hypertension. 1994;23:857–860. doi: 10.1161/01.hyp.23.6.857. [DOI] [PubMed] [Google Scholar]
- 26.Okamura T, Okunishi H, Ayajiki K, Toda N. Conversion of angiotensin I to angiotensin II in dog isolated renal artery: role of two different angiotensin II-generating enzymes. J Cardiovasc Pharmacol. 1990;15:353–359. doi: 10.1097/00005344-199003000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Takai S, et al. Characterization of chymase from human vascular tissues. Clin Chim Acta. 1997;265:13–20. doi: 10.1016/s0009-8981(97)00114-9. [DOI] [PubMed] [Google Scholar]
- 28.Caughey GH, Zerweck EH, Vanderslice P. Structure, chromosomal assignment, and deduced amino acid sequence of a human gene for mast cell chymase. J Biol Chem. 1991;266:12956–12963. [PubMed] [Google Scholar]
- 29.Takai S, Shiota N, Kobayashi S, Matsumura E, Miyazaki M. Induction of chymase that forms angiotensin II in the monkey atherosclerotic aorta. FEBS Lett. 1997;412:86–90. doi: 10.1016/s0014-5793(97)00752-7. [DOI] [PubMed] [Google Scholar]
- 30.Jeziorska M, McCollum C, Woolley DE. Mast cell distribution, activation, and phenotype in atherosclerotic lesions of human carotid arteries. J Pathol. 1997;182:115–122. doi: 10.1002/(SICI)1096-9896(199705)182:1<115::AID-PATH806>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Muller DN, et al. Local angiotensin II generation in the rat heart: role of renin uptake. Circ Res. 1998;82:13–20. doi: 10.1161/01.res.82.1.13. [DOI] [PubMed] [Google Scholar]
- 32.Balcells E, Meng QC, Johnson WJ, Oparil S, Dell’Italia LJ. Angiotensin II formation from ACE and chymase in human and animal hearts: methods and species considerations. Am J Physiol. 1997;273:H1769–H1774. doi: 10.1152/ajpheart.1997.273.4.H1769. [DOI] [PubMed] [Google Scholar]
- 33.Leife R, Estevao R, Resende AC, Salgado MC. Role of endothelium in angiotensin II formation by the rat aorta and arterial bed. Braz J Med Biol Res. 1997;30:649–656. doi: 10.1590/s0100-879x1997000500013. [DOI] [PubMed] [Google Scholar]
- 34.Juul B, Aalkjaer C, Mulvany MJ. Contractile effects of tetradecapeptide renin substrate on rat femoral resistance vessels. J Hypertens Suppl. 1987;5:S7–S10. doi: 10.1097/00004872-198707002-00002. [DOI] [PubMed] [Google Scholar]
- 35.Ideishi M, Sasaguri M, Ikeda M, Arakawa K. Substrate-dependent angiotensin II formation in the peripheral circulation. Life Sci. 1990;46:335–341. doi: 10.1016/0024-3205(90)90012-g. [DOI] [PubMed] [Google Scholar]
- 36.Andre P, Schott C, Nehlig H, Stoclet JC. Aortic smooth muscle cells are able to convert angiotensin I to angiotensin II. Biochem Biophys Res Commun. 1990;173:1137–1142. doi: 10.1016/s0006-291x(05)80904-0. [DOI] [PubMed] [Google Scholar]
- 37.Ideishi M, Noda K, Sasaguri M, Ikeda M, Arakawa K. Angiotensin II forming activity of vascular endothelial and smooth muscle cells. Artery. 1993;20:95–102. [PubMed] [Google Scholar]
- 38.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullenbach R, Lagoda PJ, Welter C. An efficient salt-chloroform extraction of DNA from blood and tissues. Trends Genet. 1989;5:391. [PubMed] [Google Scholar]
- 40.Kozak M. An analysis of 5_-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowan KN, Jones PL, Rabinovitch M. Elastase and matrix metalloproteinase inhibitors induce regression, and tenascin-C antisense prevents progression, of vascular disease. J Clin Invest. 2000;105:21–34. doi: 10.1172/JCI6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson IA, et al. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 43.Urata H, Karnik SS, Graham RM, Husain A. Dipeptide processing activates recombinant human prochymase. J Biol Chem. 1993;268:24318–24322. [PubMed] [Google Scholar]
- 44.Kido H, Fukuseen N, Katunuma N. Chymotrypsin- and trypsin-type serine proteases in rat mast cells: properties and functions. Arch Biochem Biophys. 1985;239:436–443. doi: 10.1016/0003-9861(85)90709-x. [DOI] [PubMed] [Google Scholar]
- 45.Benfey PN, Yin FH, Leder P. Cloning of the mast cell protease, RMCP II. Evidence for cell-specific expression and a multi-gene family. J Biol Chem. 1987;262:5377–5384. [PubMed] [Google Scholar]
- 46.Takahashi H, Nukiwa T, Basset P, Crystal RG. Myelomonocytic cell lineage expression of the neutrophil elastase gene. J Biol Chem. 1988;263:2543–2547. [PubMed] [Google Scholar]
- 47.Kovanen PT. Role of mast cells in atherosclerosis. Chem Immunol. 1995;62:132–170. [PubMed] [Google Scholar]
- 48.Wypij DM, et al. Role of mast cell chymase in the extracellular processing of big-endothelin-1 to endothelin-1 in the perfused rat lung. Biochem Pharmacol. 1992;43:845–853. doi: 10.1016/0006-2952(92)90252-e. [DOI] [PubMed] [Google Scholar]
- 49.Reilly CF, Schechter NB, Travis J. Inactivation of bradykinin and kallidin by cathepsin G and mast cell chymase. Biochem Biophys Res Commun. 1985;127:443–449. doi: 10.1016/s0006-291x(85)80180-7. [DOI] [PubMed] [Google Scholar]
- 50.Caughey GH, Leidig F, Viro NF, Nadel JA. Substance P and vasoactive intestinal peptide degradation by mast cell tryptase and chymase. J Pharmacol Exp Ther. 1988;244:133–137. [PubMed] [Google Scholar]
- 51.Fang KC, Raymond WW, Lazarus SC, Caughey GH. Dog mastocytoma cells secrete a 92-kD gelatinase activated extracellularly by mast cell chymase. J Clin Invest. 1996;97:1589–1596. doi: 10.1172/JCI118583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang KC, Raymond WW, Blount JL, Caughey GH. Dog mast cell alpha-chymase activates progelatinase B by cleaving the Phe88-Gln89 and Phe91-Glu92 bonds of the catalytic domain. J Biol Chem. 1997;272:25628–25635. doi: 10.1074/jbc.272.41.25628. [DOI] [PubMed] [Google Scholar]
- 53.Vartio T, Seppa H, Vaheri A. Susceptibility of soluble and matrix fibronectins to degradation by tissue proteinases, mast cell chymase and cathepsin G. J Biol Chem. 1981;256:471–477. [PubMed] [Google Scholar]
- 54.Sage H, Woodbury RG, Bornstein P. Structural studies on human type IV collagen. J Biol Chem. 1979;254:9893–9900. [PubMed] [Google Scholar]
- 55.Reilly CF, Tewksbury DA, Schechter NM, Travis J. Rapid conversion of angiotensin I to angiotensin II by neutrophil and mast cell proteinases. J Biol Chem. 1982;257:8619–8622. [PubMed] [Google Scholar]
- 56.Sanker S, et al. Distinct multisite synergistic interactions determine substrate specificities of human chymase and rat chymase-1 for angiotensin II formation and degradation. J Biol Chem. 1997;272:2963–2968. doi: 10.1074/jbc.272.5.2963. [DOI] [PubMed] [Google Scholar]
- 57.Chandrasekharan UM, Sanker S, Glynias MJ, Karnik SS, Husain A. Angiotensin II-forming activity in a reconstructed ancestral chymase. Science. 1996;271:502–505. doi: 10.1126/science.271.5248.502. [DOI] [PubMed] [Google Scholar]
- 58.McGuire MJ, Lipsky PE, Thiele DL. Generation of active myeloid and lymphoid granule serine proteases requires processing by the granule thiol protease dipeptidyl peptidase I. J Biol Chem. 1993;268:2458–2467. [PubMed] [Google Scholar]
- 59.Dikov MM, Springman EB, Yeola S, Serafin WE. Processing of procarboxypeptidase A and other zymogens in murine mast cells. J Biol Chem. 1994;269:25897–25904. [PubMed] [Google Scholar]
- 60.Murakami M, Karnik SS, Husain A. Human prochymase activation. A novel role for heparin in zymogen processing. J Biol Chem. 1995;270:2218–2223. [PubMed] [Google Scholar]
- 61.Mizuno K, Tani M, Niimura S, Fukuchi S. Effect of delapril on the vascular angiotensin II release in isolated hind legs of the spontaneously hypertensive rat: evidence for potential relevance of vascular angiotensin II to the maintenance of hypertension. Clin Exp Pharmacol Physiol. 1991;18:619–625. doi: 10.1111/j.1440-1681.1991.tb01635.x. [DOI] [PubMed] [Google Scholar]
- 62.Nakano N, et al. Role of angiotensin II in the regulation of a novel vascular modulator, hepatocyte growth factor (HGF), in experimental hypertensive rats. Hypertension. 1997;30:1448–1454. doi: 10.1161/01.hyp.30.6.1448. [DOI] [PubMed] [Google Scholar]
- 63.Place GA, Chetland J, Galpin IJ, Benyon RJ. The effect of analogues of chymostatin on lysosomal and non-lysosomal components of protein degradation in isolated hepatocytes. Biochim Biophys Acta. 1987;925:185–193. doi: 10.1016/0304-4165(87)90108-5. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt M, Loffler B-M, Kuntze H. Protease inhibitors reduce lysosomal acid phospholipase A1 activity in cultured rat hepatocytes. Biochem Int. 1990;20:337–342. [PubMed] [Google Scholar]
- 65.Billings PC, Newberne PM, Kennedy AR. Protease inhibitor suppression of colon and anal gland carcinogenesis induced by dimethylhydrazine. Carcinogenesis. 1990;11:1083–1086. doi: 10.1093/carcin/11.7.1083. [DOI] [PubMed] [Google Scholar]
- 66.Yavelow J, Caggana M, Beck KA. Proteases occurring in the cell membrane: a possible cell receptor for the Bowman-Birk type of protease inhibitors. Cancer Res. 1987;47:1598–1601. [PubMed] [Google Scholar]
- 67.De Mello WC, Danser AH. Angiotensin II and the heart: on the intracrine renin-angiotensin system. Hypertension. 2000;35:1183–1188. doi: 10.1161/01.hyp.35.6.1183. [DOI] [PubMed] [Google Scholar]
- 68.Sen I, Rajasekaran AK. Angiotensin II-binding protein in adult and neonatal rat heart. J Mol Cell Cardiol. 1991;23:563–572. doi: 10.1016/0022-2828(91)90048-q. [DOI] [PubMed] [Google Scholar]
- 69.Fu ML, et al. Immunohistochemical localization of angiotensin II receptors (AT1) in the heart with anti-peptide antibodies showing a positive chronotropic effect. Receptors Channels. 1998;6:99–111. [PubMed] [Google Scholar]
- 70.Sadoshima J, Xu Y, Slayter HS, Izumo S. Autocrine release of angiotensin II mediates stretch-induced hypertrophy of cardiac myocytes in vitro. Cell. 1993;75:977–984. doi: 10.1016/0092-8674(93)90541-w. [DOI] [PubMed] [Google Scholar]
- 71.Aharinejad S, et al. Spontaneously hypertensive rats develop pulmonary hypertension and hypertrophy of pulmonary venous sphincters. Am J Pathol. 1996;148:281–290. [PMC free article] [PubMed] [Google Scholar]
- 72.Lutzelschwab C, Pejler G, Aveskogh M, Hellman L. Secretory granule proteases in rat mast cells. Cloning of 10 different serine proteases and a carboxypeptidase A from various rat mast cell populations. J Exp Med. 1997;185:13–29. doi: 10.1084/jem.185.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]