Abstract
Congenital erythropoietic porphyria, an autosomal recessive inborn error of heme biosynthesis, results from the markedly deficient activity of uroporphyrinogen III synthase. Extensive mutation analyses of 40 unrelated patients only identified approximately 90% of mutant alleles. Sequencing the recently discovered erythroid-specific promoter in six patients with a single undefined allele identified four novel mutations clustered in a 20-bp region: (a) a –70T to C transition in a putative GATA-1 consensus binding element, (b) a –76G to A transition, (c) a –86C to A transversion in three unrelated patients, and (d) a –90C to A transversion in a putative CP2 binding motif. Also, a –224T to C polymorphism was present in approximately 4% of 200 unrelated Caucasian alleles. We inserted these mutant sequences into luciferase reporter constructs. When transfected into K562 erythroid cells, these constructs yielded 3 ± 1, 54 ± 3, 43 ± 6, and 8 ± 1%, respectively, of the reporter activity conferred by the wild-type promoter. Electrophoretic mobility shift assays indicated that the –70C mutation altered GATA1 binding, whereas the adjacent –76A mutation did not. Similarly, the –90C mutation altered CP2 binding, whereas the –86A mutation did not. Thus, these four pathogenic erythroid promoter mutations impaired erythroid-specific transcription, caused CEP, and identified functionally important GATA1 and CP2 transcriptional binding elements for erythroid-specific heme biosynthesis.
Introduction
Congenital erythropoietic porphyria (CEP), also known as Günther disease, is an autosomal recessive inborn error that results from the markedly deficient, but not absent, activity of the fourth enzyme in the heme biosynthetic pathway, uroporphyrinogen III synthase (URO-synthase; EC 4.2.1.75; hydroxymethylbilane hydrolase [cyclizing]) (1). The resultant accumulation of the nonphysiological porphyrin isomer uroporphyrin I (URO I) in erythrocytes leads to hemolysis, and the released porphyrin isomer is deposited in tissues and bones and is excreted in the urine and feces. Ultraviolet light activates the phototoxic URO I, resulting in tissue damage and the formation of bullous lesions that rupture and often become infected, leading to cutaneous scarring, bone resorption, and deformities (2). The clinical manifestations are markedly heterogeneous, ranging from nonimmune hydrops fetalis due to severe hemolytic anemia in utero to milder, later-onset forms, which have only cutaneous photosensitivity in adult life (3–6). Severely affected patients are transfusion-dependent throughout life, have secondary hypersplenism and are usually disfigured. The only effective treatment for severely affected patients has been bone marrow transplantation (see, for example, ref. 7).
Human URO-synthase from erythrocytes has been purified to homogeneity and shown to be a monomeric protein with an apparent molecular mass of 29.5 kDa (8). The full-length cDNA encoding the human URO-synthase polypeptide of 265 amino acids was isolated, sequenced, and expressed in Escherichia coli (9). Chromosome mapping localized a single human URO-synthase gene to the region, l0q25.3→q26.3 (10). The availability of the URO-synthase cDNA (9) and recently published (11) intron/exon boundaries enabled investigation of the molecular lesions causing CEP. To date, 22 mutations causing CEP have been described in the URO-synthase gene, including single base substitutions, insertions, deletions, and splicing defects (12). Most mutations have been identified in one or a few unrelated families with the exception of the severe C73R lesion that occurred in about 30% of mutant alleles studied (2). Prokaryotic expression of the URO-synthase missense mutations identified those with residual activity, thereby enabling genotype/phenotype predictions for mild to severe disease manifestations (2, 13–15). However, extensive molecular analysis of the coding and adjacent flanking, intronic, and 5′- and 3′-untranslated sequences did not detect the causative mutations in about 10–15% of mutant alleles from unrelated patients (2, 15, 16). In our series of 40 unrelated patients with CEP, both URO-synthase mutant alleles were identified in each patient, except in 11 in whom only one mutation was detected (11 unidentified mutations in 80 alleles, or 13.8%) (2).
Recently, the approximately 34-kb human gene encoding URO-synthase was isolated and characterized (11). Relevant to these studies, the gene had alternative promoters that generated erythroid-specific and housekeeping transcripts, suggesting the possibility that mutations in the erythroid promoter could cause CEP. In this communication, we report a cluster of four pathogenic mutations in a 20-bp region of the URO-synthase erythroid-specific promoter. These mutations were identified in six patients with CEP, one at –70 altering a GATA1 consensus site, one at –76 adjacent to the GATA1 site, as well as three at –86 and one at –90, both occurring in a putative CP2 binding site. Luciferase promoter/reporter gene analyses demonstrated that the –70C and –90A mutations severely impaired promoter function, whereas the –76A and –86A substitutions decreased reporter gene activity about 45% and 55%, respectively. Electrophoretic mobility shift assays (EMSAs) indicated that the –70C lesion impaired GATA1 binding, and the –90A mutation impaired CP2 binding, whereas the –76A mutation had no effect on GATA1 binding. The –86A mutation appeared to enhance CP2 binding. These results demonstrate the occurrence of functionally important GATA1 and CP2 binding sites for the erythroid-specific transcription of human URO-synthase.
Methods
Patients specimens and disease severity classification.
Peripheral blood samples were collected with informed consent from patients with CEP with one unidentified mutation and from their family members. Lymphoid cell lines were established using cyclosporin A and Epstein-Barr virus as described previously (17). Cells were maintained by standard procedures in RPMI 1640 media (Life Technologies Inc., Gaithersburg, Maryland, USA). Peripheral blood samples were also obtained with informed consent from 70 normal Caucasian individuals and 30 other, unrelated CEP patients, for restriction analysis to determine whether the promoter mutations were disease-causing or silent polymorphisms. For phenotyping, the following clinical criteria were used. Patients who developed nonimmune hydrops fetalis and/or were transfusion-dependent at any age were classified as severe. Transfusion-independent patients were classified as moderate or mild depending on their age, hematologic indices, splenic size, and/or extent and severity of their cutaneous lesions.
Detection of URO-synthase exonic and flanking mutations.
For each of the unrelated patients with a previously undefined mutation, efforts were undertaken to resequence their URO-synthase alleles. Genomic DNA was extracted from lymphoblasts using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minnesota, USA) and each exon, including its intron/exon boundaries, was amplified by PCR using primer sets in which one primer was biotinylated as described previously (14).
Detection of URO-synthase erythroid promoter mutations.
To identify possible mutations in the erythroid promoter, a region of 840 bp, which included 53 bp of exon 2A and the adjacent upstream 787 bp of intron 1 (GenBank Accession Numbers AF230663 and AF230664; ref. 11), was PCR-amplified using sense (no. 2 biotinylated) and antisense (no. 3) primers (Table 1). Each 100 μl amplification reaction contained 2 μg of genomic DNA, 100 pmol of each primer, 10 nmol of each dNTP, 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton x100, and 2 U of Taq polymerase (Promega Corp., Madison, Wisconsin, USA). After an initial 5-minute incubation at 94°C, amplification (35 cycles) was performed with denaturation at 94 °C for 1 minute, annealing at 65°C for 1 minute, and extension at 72°C for 1 minute. A further 10-minute extension at 72°C was carried out after completion of the 35 cycles. The PCR products were purified by the Wizard PCR Prep DNA Purification System (Promega Corp.). An aliquot of the purified PCR product (40–80 μl) was denatured, and the biotinylated single strands were isolated by affinity capture using streptavidin-coated paramagnetic beads (Dynal Inc., Great Neck, New York, USA) (14). The pelleted beads were resuspended in 7 μl of distilled water and sequenced by the dideoxy chain termination method (18) using Sequenase according to the manufacturer’s instructions (United States Biochemical Corp., Cleveland, Ohio, USA). The sequencing primers are shown in Table 1. The sequenced products were loaded on 4% acrylamide gels (19:1) containing 7 M urea. The gels were dried, exposed to Kodak XOMAT film (Eastman Kodak Co., Rochester, New York, USA) for 1–2 days, and then developed. Alternatively, the PCR products were sequenced using an ABI Prism 377 DNA Sequencer and the ABT Prism BigDye Terminator Ready Reaction mix (Applied Biosystems, Foster City, California, USA).
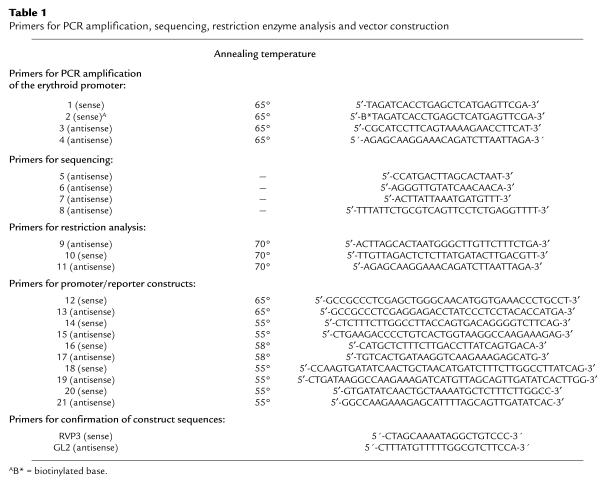
Table 1.
Primers for PCR amplification, sequencing, restriction enzyme analysis and vector construction
Confirmation of the erythroid promoter base substitutions by restriction analysis.
Computer-assisted analyses revealed that all of the mutations and the polymorphism modified restriction sites. For the following restriction analyses of the erythroid promoter, genomic DNA was amplified using the conditions described above with the primers specified in Table 1. Restriction enzymes and molecular weight standards were purchased from New England Biolabs Inc. (Beverly, Massachusetts, USA). The PCR products were purified by the Wizard PCR Prep DNA Purification System. For the –70C and –76A mutations, genomic DNA was amplified using primers 10 and 11, respectively. To detect the –86A mutation, genomic DNA was amplified using sense and antisense primers 1 and 4, respectively. For the –90A mutation or the –224C substitution, genomic DNA was amplified using sense and antisense primers 9 and 10, respectively.
Haplotyping of the –86A mutant alleles.
To determine whether the three probands who had the –86A mutation were related, haplotyping was performed on genomic DNAs from appropriate family members. Four CA repeat markers flanking the URO-synthase gene at 10q26.1 were obtained from Research Genetics (Huntsville, Alabama, USA). The fluorescent-labeled markers, from centromere to telomer, were D10S209, D10S2322, D10S216, and D10S575 (19). PCR amplification and gel analysis were performed as described previously (20, 21).
Construction of the URO-synthase promoter/reporter plasmids.
To assess the effects of the erythroid promoter mutations on promoter function, a series of promoter/enhancer reporter constructs were made in the pGL3-Basic Luciferase Reporter Vector (Promega Corp.), which lacks eukaryotic promoter and enhancer sequences, and was designed to evaluate putative promoter sequences. To construct the wild-type erythroid promoter/reporter plasmid, designed hEPr-1, 647 bp of the wild-type URO-synthase erythroid promoter region upstream of exon 2A from –1 to –647 was PCR amplified from genomic DNA using sense and antisense primers 12 and 13, respectively. For cloning purposes, 5′-tails containing an Xho I restriction site were included in the sense and antisense primers, respectively, preceded by a GCCGCC clamp sequence (Table 1). After amplification, the PCR products were digested with Xho I and subcloned into the corresponding sites of the pGL3-Basic Luciferase Reporter Vector. The plasmid was purified using the Qiagen Plasmid Maxi Kit (QIAGEN Inc., Valencia, California, USA), and the authenticity of the wild-type pGL3-construct was determined by automated sequencing with primers RVP3 and GL2, located in the pGL3 vector, using an ABI PRISM, Model 377 sequencer (Applied Biosystems). To construct promoter/reporter plasmids for the –70C, –76A, –86A, and –90A mutations, the wild-type hEPr-1 (promoter/reporter pGL3) construct was used as parental DNA for performing mutagenesis using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, California, USA). The sense and antisense primers used to introduce the –70C, –76A, –86A, and –90A mutations were 14 and 15, 16 and 17, 18 and 19, and 20 and 21, respectively (Table 1). Two additional constructs were made to create reporter constructs containing –80C→A and –110C→A mutations in putative CP2 elements. The mutagenesis procedure was performed according to the manufacturer’s instructions. The authenticity of the mutant pGL3-constructs was determined by automated sequencing using primers RVP3 and GL2 listed in Table 1.
Cell culture, transfections, and luciferase assays.
Human K562 erythroleukemia cells were grown in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin (Life Technologies Inc.). For transfection, 8 × 105 K562 cells were transfected with 2 μg of each promoter/reporter construct and 50 ng of the internal control Renilla Luciferase-TK (pRL-TK) vector, using 5 μl of DMRIE-C reagent (Life Technologies Inc.). After each transfection (48 hours), the cells were collected by centrifugation, washed twice with PBS, and lysed with 50 μl of Passive Lysis Buffer (Promega Corp.). To induce the K562 cells, hemin was added to the media to a final concentration of 25 μM 24 hours before transfection. The Renilla and firefly luciferase activities were determined in 20 μl of the cell lysates using the Dual-Luciferase Reporter Assay System (Promega Corp.) according to the manufacturer’s instructions, with a Microtiter Plate Luminometer (Model MLX; DYNEX Technologies Inc., Chantilly, Virginia, USA). The firefly luciferase activities were normalized for transfection efficiency using the Renilla luciferase activity as an internal control. The data were expressed as a percentage of the activity of the test promoter/reporter construct and that obtained for the wild-type erythroid promoter/reporter construct, hEPr-1 under the same conditions. A negative control using the pGL-3 Basic Vector was included in all experiments. The results are reported as the means ± SD of at least four independent transfection experiments.
EMSAs.
The binding reactions were performed using the GATA-1 GelshiftKit (Geneka Biotechnology Inc., Montreal, Quebec, Canada) according to the manufacturer’s instructions. K562 nuclear extracts were obtained from Geneka. The oligonucleotide probes were synthesized using an ABI Model 394 DNA/RNA Synthesizer (Applied Biosystems). The upper strand of each oligonucleotide is shown in Table 1. An oligonucleotide probe containing a GATA1 consensus binding site from the SCL (stem-cell leukemia hematopoietic transcription factor) gene promoter was obtained from Geneka, and the murine α-globin probe for CP2 (GenBank Acc. no. X05379) was synthesized as described above. Double-stranded oligonucleotides were labeled with [γ-32P]-ATP (NEN Life Science Products Inc., Boston, Massachusetts, USA) using T4 DNA polynucleotide kinase (New England Biolabs). Specificity of the binding reactions was determined by incubation of the probe and nuclear extract in the presence of 100-fold molar excess of the indicated competitor oligonucleotides. After a 20-minute preincubation of nuclear extract with gel-shift buffers, the radiolabeled probe (0.5 ng per reaction) and any competitor were incubated with the nuclear extract mixture for 20 minutes at 4°C. Supershift reactions were conducted by adding preimmune serum or specific antibodies plus Protein A to the binding reaction mixture, following the second 20-minute incubation. Rat monoclonal anti-GATA1 antibody was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, California, USA), and rabbit polyclonal anti-CP2 (anti–LBP-1) was kindly supplied by R. Roeder (Rockefeller University, New York, New York, USA). Staphylococcus aureus Protein A was obtained from Sigma (St. Louis, Missouri, USA). The immune precipitation reactions were carried out at 4°C for 60 minutes. The protein-DNA complexes were analyzed by electrophoresis on nondenaturing 5% polyacrylamide gels.
Results
Identification of the erythroid promoter URO-synthase mutations.
Five base substitutions were detected by sequencing the URO-synthase erythroid promoter from the 11 patients with CEP in which only one mutant allele had been previously identified. These included: (a) a T→C transition at nucleotide (nt) –70 in proband 1, a male fetus with nonimmune hydrops fetalis, (b) a G→A transition at nt –76 in proband 2, a male with mild cutaneous disease, (c) a C→A transversion at nt –86 in probands 3 to 5, unrelated adults with mild cutaneous disease, (d) a C→A transversion at nt –90 in proband 6, an adult male with a moderately severe phenotype, and (e) a T→C transition at nt –224 that was detected in proband 1. These lesions were confirmed by sequencing genomic DNA from parents (probands 1 and 3) and restriction analyses of genomic DNA from all probands and the relatives of probands 1 and 3 to 6. In each case, amplification and sequencing of genomic DNA from the parents of the CEP probands demonstrated the presence of the mutant allele in a parent with the exception of proband 1’s healthy father, who was heterozygous for mutation –70C and homozygous for the –224C lesion.
Restriction analyses also confirmed the presence of the –70C, –76A, –86A, –90A, and –224C base substitutions in the probands and their respective relatives. The –70C mutation created a Bsr I site, and the 475-bp PCR product containing the –70C lesion generated 306- and 169-bp fragments, whereas the normal allele (–70T) was not digested. The –76A mutation abolished a Hae III site, and the 475-bp PCR product containing the mutation was not digested, whereas the other allele was cleaved, resulting in 300- and 175-bp fragments. The –86A mutation created a Sau 3AI site and digestion of the 780-bp PCR product from probands 2 to 5 revealed 593-, 158-, 18-, and 11-bp fragments, whereas the other allele generated a 751-bp fragment containing the –86C site and the 18- and 11-bp fragments. The –90A lesion abolished a Nla III site and the 351 PCR-product was not digested, whereas the other allele was cleaved, resulting in 289- and 62-bp fragments. The –224C base substitution abolished an Eco RV restriction site, and the 351 bp PCR-product containing the lesion was cleaved into 273- and 78-bp fragments, whereas the –224T containing amplicon was digested into 149-, 124-, and 78-bp fragments. In addition, these restriction analyses were performed on genomic DNAs from 70 normal Caucasian individuals (140 alleles) and 30 other CEP homozygotes in whom both mutant alleles were known (60 alleles). These studies did not identify –70C, –76A, –86A, or –90A mutations in any normal individuals or in patients with CEP with two known mutations. The –224C lesion was detected in eight of the 160 normal alleles and in none of the other CEP alleles tested, indicating that the –224T to C is a rare polymorphism. As already noted here, the only patient with CEP with the –224C polymorphism was proband 1 whose phenotypically normal father was a –224C/–224C homozygote, providing further evidence that this lesion is a neutral polymorphism.
Haplotyping analysis of patients with CEP with the –86C mutation.
Given that three apparently unrelated patients with CEP had the –86A mutation, haplotype analysis was performed using flanking microsatellite markers that spanned a region of about 5 cM proximal and 0.2 cM distal to the URO-synthase gene, respectively. The haplotypes of all three patients with the –86A mutation were different at the four closely linked CA repeat markers (Table 2), indicating that the three –86A mutations resulted from independent mutational events. Thus, a total of six independent mutations occurred in this 20-bp region.
Table 2.
Haplotype analysis of patients with CEP with the –86A mutation
Erythroid promoter mutations impair transcription.
To assess the functionality of the erythroid promoter mutations, luciferase promoter/reporter gene constructs were prepared, and their respective activities in transfected K562 cells were compared to that of the wild-type promoter/reporter construct containing 647 bp of the erythroid promoter upstream of exon 2A. Table 3 shows the mean luciferase activities from at least three independent transfections for the –70C, –76A, –86A, and –90A mutant promoter/reporter constructs 48 hours after transfection into uninduced or hemin-induced K562 erythroleukemia cells. The mean luciferase activities of the –70C and –90A promoter/reporter constructs in uninduced K562 cells were approximately 3% and 8%, respectively, of the activity generated by the wild-type erythroid promoter/reporter construct, whereas the –76A and –86A mutant constructs had about 54% and 43% of wild-type activity, respectively. The luciferase activities of the mutant constructs were similarly reduced in K562 cells induced to differentiate with hemin.
Table 3.
Luciferase activity of wild-type and mutant promoter/reporter constructs in uninduced and induced K652 cells
Reporter constructs containing mutations of the conserved cytosines at –80C→A and –110C→A in the CP2 elements had mean luciferase activities of 21% (n = 3) and 93% (n = 3) of wild-type, respectively. These findings indicated that the CP2 site at –80 was functional, whereas the upstream CP2 core motif at –110 was not, further indicating that only the CP2 elements adjacent to the GATA1 site at –70 were functional.
EMSAs.
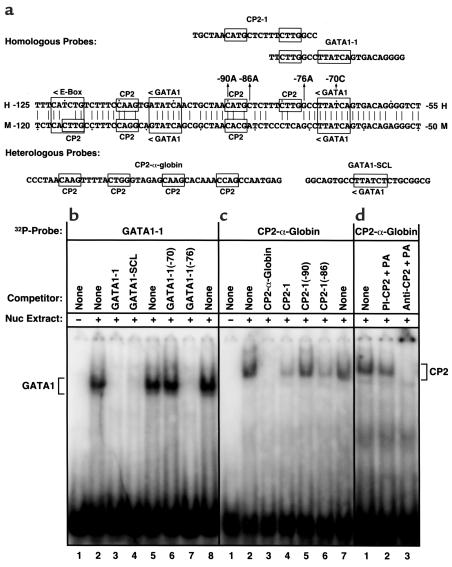
Because mutations occurred in putative GATA1 and CP2 transcriptional binding sites, EMSAs were performed to evaluate the effect of these mutations on GATA1 and CP2 binding. Figure 1a shows the erythroid promoter region containing putative GATA1, CP2, and E-box binding elements and indicates the probes used for EMSAs that were designed to evaluate GATA1 and/or CP2 binding. Figure 1, b and c, show that the radiolabeled wild-type GATA1 and CP2 (α-globin sequence) oligonucleotide probes bound proteins that were present in K562 nuclear extracts. The binding activity of the GATA1 wild-type URO-synthase oligonucleotide probe (GATA1-1) was inhibited when incubated with 100-fold mass excess of either the wild-type probe (GATA1-1) or a known heterologous GATA1 oligonucleotide probe SCL (22) as cold competitors (Figure 1b, lanes 3 and 4). Competition studies with cold oligonucleotide probes with the –70C or –76A mutations were performed to determine whether these mutations altered GATA1 binding activity. No competition was observed with the –70C oligonucleotide probe, indicating that this mutation totally abolished GATA1 binding activity. In contrast, the –76A oligonucleotide completely competed GATA1 binding, demonstrating that the –76A mutation did not alter GATA1 binding to this probe.
Figure 1.
EMSAs of GATA1 and CP2 binding by the URO-synthase erythroid promoter and the α-globin promoter, respectively. EMSAs were performed as described in Methods. (a) Partial sequence of the human and murine (36) URO-synthase erythroid-specific promoters. The location and orientation (<, >) of the GATA1, E-box, and CP2 erythroid binding elements are indicated, as are the four novel promoter mutations. Dots are placed every tenth nucleotide. The heterologous CP2 site from the murine α-globin gene promoter and the GATA1 site from the human SCL gene promoter are shown. (b) The radiolabeled probe is GATA1-1 (see a). K562 nuclear extract was present in lanes 2–8. A 100-fold mass excess of unlabeled competitor is included in the following lanes: lane 3, GATA1-1; lane 4, GATA1-SCL; lane 6, GATA1-1, –70 mutation; lane 7, GATA1-1, –76 mutation. (c) The radiolabeled probe is CP2-α-globin (see a). K562 nuclear extract was present in lanes 2–6. Unlabeled competitor is included in the following lanes: lane 3, CP2-α-globin; lane 4, CP2-1; lane 5, CP2-1(–90); lane 6, CP2-1(–86). (d) The radiolabeled probe was CP2-α-globin (see a). K562 nuclear extract was present in lanes 1 to 3. Lane 2: incubation with preimmune serum and Protein A. Lane 3: incubation with anti-CP2 antibody and Protein A.
EMSA of the putative CP2 binding site in the human URO-synthase erythroid promoter was assessed using the prototype 47-bp CP2 binding sequence from the murine α-globin promoter. This probe (CP2-α-globin) formed a complex with a protein(s) in K562 nuclear extracts (Figure 1c, lane 2). This binding activity was completely competed by the wild-type cold CP2-α-globin probe (lane 3) and strongly competed by the wild-type 23-bp CP2 probe, CP2-1 (lane 4), and 23-bp CP2-1 URO-synthase promoter probe containing the –86C→A mutation (lane 6). In contrast, the CP2-1 probe containing the –90A mutation failed to compete CP2 protein binding activity (lane 5), indicating that the –90A mutation markedly, if not completely, abolished CP2 binding to this region of the URO-synthase gene. Supershift analysis with a polyclonal anti–α-CP2 antibody plus Protein A completely shifted the binding activity to the top of the gel, whereas the preimmune antisera had no effect. Although the 100-fold excess homologous URO-synthase CP2-1 probe displaced bona fide CP2 from the heterologous α−globin probe, no shifted band was detected with radiolabeled CP2-1, indicating weak binding to CP2 or a CP2-like protein. Thus, these results indicated that the GATA1 and CP2 elements in the –70 to –90 bp region were transcriptionally functional and that mutations in critical residues of either binding motif resulted in severe impairment of transcriptional activity.
Genotype/phenotype studies.
Table 4 indicates the genotypes, phenotypes, and relative in vitro activities of the respective erythroid promoter and coding region mutations for the six CEP probands. Proband 1 who expired in utero from nonimmune hydrops fetalis was heteroallelic for the –70C erythroid promoter mutation that markedly impaired transcription (<3% wild-type luciferase activity) and the severe C73R mutation that had little, if any, detectable URO-synthase activity when expressed in E. coli (23). Probands 2, 3, 4, and 5 had normal hemoglobin values or very mild anemia and only mild cutaneous photosensitivity, which was exacerbated when exposed to sunlight. The residual activity expressed by the –76A and –86A alleles (∼54% and ∼43% of wild-type luciferase activity, respectively) produced sufficient enzyme activity even in the presence of their more severe alleles (C73R, C73R, IVS2+1, and 398insG, respectively) which produced little or no functional enzyme. Proband 6 had a severe phenotype with chronic anemia, transfusion dependency, and cutaneous photosensitivity, consistent with his genotype, the severe –90C mutation that markedly impaired transcription (<8% of wild-type luciferase activity) and the severe G225S missense mutation that expressed less than 2% of normal enzyme activity in vitro.
Table 4.
Genotype-phenotype correlations in CEP probands with erythroid promoter mutations
Discussion
To date, 22 URO-synthase mutations causing CEP have been identified in the coding region or intronic flanking regions (2, 12). Only the C73R mutation was common, having been found in about 35% of the more than 120 alleles studied (2, 13, 16). Notably, about 10–15% of the mutant alleles causing CEP have not been detected by sequencing the entire coding region and adjacent intron-exon boundaries, suggesting other sites for the missing mutations (2, 15, 16). Our recent discovery that the human URO-synthase gene had alternative housekeeping and erythroid-specific promoters (11) suggested the possibility that the undetected mutations were in the erythroid promoter or its unique 5′ untranslated sequence. The finding of four erythroid promoter mutations confirmed this notion, and identified the elusive mutations in six CEP patients in which previous mutation analysis of the exonic and adjacent flanking intronic sequences had only detected a single mutation. Of the three patients with the –86A mutation, haplotype analyses indicated that they were not related and that their mutations arose independently. Thus, six independent mutations occurred in the 20 nucleotides from –70 to –90, defining this region as a mutational hot spot. The prevalence of the four promoter mutations in our series of 40 patients with CEP is 7.5% (six of 80 mutant alleles). Although the discovery of these erythroid promoter mutations accounts for the mutations in six of the 11 patients studied, at least one, if not several other, cryptic mutations remain unidentified.
Previously, genotype/phenotype correlations in this disease were based on the expression of the human URO-synthase mutations in E. coli, which provided an in vitro estimate of their in vivo residual activities (13, 14). For example, expression of the C73R allele in E. coli resulted in the detection of less than 1% of the activity expressed by the normal allele, consistent with the severe phenotype of the C73R/C73R homozygotes who present with nonimmune hydrops fetalis or severe transfusion dependency from birth (2). Similar genotype/phenotype correlations can be made for the erythroid promoter mutations based on their luciferase promoter/reporter activities (Table 4). The patient with the –70C mutation resulting in only 3% luciferase reporter activity was heteroallelic for C73R, presented with nonimmune hydrops fetalis, and subsequently died in utero (as did the mother’s previous pregnancy), confirming the deleterious nature of this promoter mutation in vivo. The patient with the –90A/G225S genotype had severe transfusion-dependent disease, consistent with a luciferase activity of only 8% and the extremely low level of expression of the G225S allele in E. coli (∼1.2% of wild-type) (14).
In contrast, the three patients with the –86A lesion (which reduced luciferase promoter/reporter activity to about 43% of that of the wild-type promoter) all had mild disease limited to cutaneous manifestations when exposed to ultraviolet light. These three patients had aggressively avoided sunlight and had minimal scarring. The patient with the –76A mutation also had mild disease limited to cutaneous involvement. The other heteroallelic mutations in these patients were all severe and included C73R (in probands 1, 2, and 3), 398insG in exon 7 (in proband 5) that resulted in a frameshift and premature polypeptide truncation, and a splicing mutation IVS2+1 (in proband 4), which was previously shown by RT-PCR to delete exon 2 in all transcripts containing the mutation (14). Thus, the mild phenotypes of these patients whose other mutations markedly impaired enzyme synthesis or activity were consistent with the fact that the –76A and –86A mutations only reduced luciferase activity by about 45% and 55% in vitro, respectively. The residual transcriptional activity associated with the –76A and –86A mutations presumably protected these patients from severe disease manifestations.
The occurrence of these four disease-causing mutations clustered within 20 bp of the erythroid promoter suggested that this region was not only a mutational hot spot, but also was functionally important for erythroid transcription and/or differentiation. Computer-assisted analysis of the transcriptional binding sites in this region revealed the presence of exact matches of a GATA1 site (TTATCA) at –68 to –73, and an adjacent CP2 site (CATGctctttCTTG) at –77 to –90 to their respective consensus sequences (WGATAR and CNRG-N6-CNR(G/C) (24–26). An additional GATA1 site (TGATAT) with a single mismatch to the consensus sequence was at –101 to –106. GATA1 (27) is the major erythroid transcriptional factor essential for globin gene expression as well as that of most erythroid-specific genes (28, 29) and forms transcriptional complexes with several erythroid transcriptional factors including “friend of GATA1” (FOG), EKLF, TAL1, and LMO2 (30–32). CP2, also known as LBP1 or LSF, is a member of a family of transcription factors, involved in the regulation of nonerythroid genes (33, 34) as well as the murine α-globin gene (24) and the inducible expression of the human γ-globin gene in thalassemia patients treated with butyrate (35). CP2 has a binding sequence that involves two CNRG motifs separated by six nucleotides; its transcription factor functions as a homo- or heterodimer (24) and has been shown to be involved in complexes with various transcriptional factors including NF-E4 and Fe65 (34, 36), but not previously with GATA1. Of note, the human GATA1 and CP2 transcriptional binding sites were conserved in position and sequence in the murine URO-synthase promoter (Figure 1a) (37). The –70C mutation altered one of the four critical core nucleotides of the GATA1 binding site, whereas the –76A mutation altered a conserved residue adjacent to the GATA1 site in an extended region of perfect homology between the human and murine sequences. The –90C mutation altered a core residue in a CP2 motif and the –86A mutation was immediately downstream, just outside of the well-conserved CP2 core sequence.
The effect of each of these novel mutations on transcription was assessed in vitro, using a luciferase promoter/reporter system in uninduced and induced K562 erythroleukemia cells. The luciferase activity generated by the –70C construct was severely reduced (<3% of wild-type activity) in both uninduced and induced K562 cells (Table 3), indicating the importance of this GATA1 site for erythroid transcription and differentiation. The –90C lesion, which was located exactly two helical turns upstream on the same DNA face as the –70 mutation, also markedly reduced luciferase activity (∼8% of wild-type) in uninduced and induced K562 cells, indicating its functional importance. The fact that the –70 and –90 mutations had a dramatic effect on promoter function suggested that these lesions may alter critical binding residues of interdependent members of an essential erythroid transcriptional complex. In contrast, the –76A and –86A promoter/reporter constructs retained about 45–55% of wild-type activity, consistent with the fact that both lesions occurred in or near a critical binding region, but did not alter essential binding nucleotides. In addition, luciferase constructs with mutations in the putative CP2 motifs at –80, –90, and –110 demonstrated the functionality of the paired –80/–90 CP2 sites (as they had only ∼21% and 8% of wild-type activity, respectively) and the lack of transcriptional function for the putative –110 CP2 motif.
To demonstrate further the function of the GATA1 and CP2 sites, EMSAs were performed with nuclear extracts from the human K562 erythroid cell line. Using the region of the GATA1 site as the radiolabeled oligonucleotide probe (i.e., GATA1-1), a specific nuclear binding complex was demonstrated that was competed by the homologous GATA1 sequence and by a heterologous human GATA1 sequence derived from the promoter of the SCL transcription factor gene. However, this binding complex was not competed by the homologous GATA1 probe containing the –70C mutation, thereby confirming that this mutation markedly impaired GATA1 binding. Analogously, EMSA studies and supershift assays demonstrated that the CP2 nuclear complex in K562 cells detected with the heterologous α-globin CP2 probe was almost completely competed by the homologous URO-synthase probe with or without the –86A mutation (Figure 1c). Notably, the URO-synthase CP2 motif containing the –90A mutation did not compete, demonstrating that this mutation markedly impaired CP2 binding. Specific binding of CP2 to the URO-synthase promoter was demonstrated by competition EMSAs; however, the radiolabeled CP2-1 probe did not detect a shift, indicating weak in vitro binding of the URO-synthase CP2-1 probe to CP2 or a CP2-like factor. In vivo, CP2 binding may require interactions with factors not present or stable in our nuclear extract and/or additional sequences in the URO-synthase gene.
Taken together, the luciferase and EMSA studies suggest that these adjacent GATA1 and CP2 binding sites are required for erythroid transcription and that mutations that disrupt or alter binding of these transcriptional factors markedly impair erythroid-specific transcription. Although GATA1 or CP2 erythroid transcriptional complexes have been described in globin and other genes (31–33), this is the first report of a transcriptional requirement for both GATA1 and CP2. Future studies should determine the role of GATA1 and CP2 in the promoter complex and the association of CP2 with transcriptional factors known to form complexes with GATA1, including FOG and TAL1/SCL.
To date, only 24 genes have promoter mutations that cause disease (11). Relatively few pathogenic regulatory lesions have been identified in housekeeping promoters, perhaps because they have evolved such that a point mutation in a single binding site usually does not markedly impair transcription (38). The only exception is the LDL-receptor gene in which a cluster of four mutations (–42 to –45) affect a Sp1 binding site (39–41). Of note, clustered point mutations in tissue-specific promoter elements have been identified in the β- and γ-globin genes, and the factor IX gene (42–47). Each of these mutation clusters occurs in a transcription factor binding site (e.g., TATA, EKLF or a CAAT box). Thus, the four promoter mutations in the URO-synthase gene described here further support the notion that clustered promoter mutations define critical elements for transcription of tissue-specific genes and that characterization of these promoter regions may identify important known or novel cis-binding elements and their respective trans-acting factors.
Acknowledgments
We thank D. Bickers, V. Cronin, N. Korman, D. M. Layton, Y. Nordmann, and C. Pierach for referral of patients; Weiming Xu for contributions to this work; and Raman Reddy (deceased) and Gregory Young for their expert technical assistance. This work was supported in part by grants from the National Institutes of Health, including a research grant (5 R01 DK26824), a grant (5 M01 RR00071) for the Mount Sinai General Clinical Research Center, and a grant (5 P30 HD28822) for the Mount Sinai Child Health Research Center. Constanza Solis is the recipient of a fellowship (96/5235) from Fondo de Investigaciones Sanitarias of Spain.
Footnotes
Constanza Solis and Gerardo I. Aizencang contributed equally to this work.
References
- 1.Anderson, K.E., Sassa, S., Bishop, D.F., and Desnick, R.J. 2001. Disorders of heme biosynthesis: X-linked sideroblastic anemia and the porphyrias. In The molecular and metabolic bases of inherited disease. C.S. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. 8th edition. McGraw-Hill. New York, New York, USA. 2961–3062.
- 2.Desnick RJ, Glass IA, Xu W, Solis C, Astrin KH. Molecular genetics of congenital erythropoietic porphyria. Semin Liver Dis. 1998;18:77–84. doi: 10.1055/s-2007-1007143. [DOI] [PubMed] [Google Scholar]
- 3.Deybach JC, et al. Congenital erythropoietic porphyria (Gunther’s disease): enzymatic studies on two cases of late onset. J Lab Clin Med. 1981;97:551–558. [PubMed] [Google Scholar]
- 4.Horiguchi Y, et al. Late onset erythropoietic porphyria. Br J Dermatol. 1989;121:255–262. doi: 10.1111/j.1365-2133.1989.tb01808.x. [DOI] [PubMed] [Google Scholar]
- 5.Verstraeten L, et al. Biochemical diagnosis of a fatal case of Gunther’s disease in a newborn with hydrops foetalis. Eur J Clin Chem Clin Biochem. 1993;31:121–128. doi: 10.1515/cclm.1993.31.3.121. [DOI] [PubMed] [Google Scholar]
- 6.Kappas, A., Sassa, S., Galbraith, R.A., and Nordmann, Y. 1995. The Porphyrias. In Metabolic and molecular bases of inherited disease. C.S. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. 7th edition. McGraw-Hill. New York, New York, USA. 2103–2160.
- 7.Tezcan I, et al. Congenital erythropoietic porphyria successfully treated by allogeneic bone marrow transplantation. Blood. 1998;92:4053–4058. [PubMed] [Google Scholar]
- 8.Tsai SF, Bishop DF, Desnick RJ. Purification and properties of uroporphyrinogen III synthase from human erythrocytes. J Biol Chem. 1987;262:1268–1273. [PubMed] [Google Scholar]
- 9.Tsai SF, Bishop DF, Desnick RJ. Human uroporphyrinogen III synthase: molecular cloning, nucleotide sequence, and expression of a full-length cDNA. Proc Natl Acad Sci USA. 1988;85:7049–7053. doi: 10.1073/pnas.85.19.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astrin KH, et al. Regional assignment of the human uroporphyrinogen III synthase (UROS) gene to chromosome 10q25.2→q26.3. Hum Genet. 1991;87:18–22. doi: 10.1007/BF01213085. [DOI] [PubMed] [Google Scholar]
- 11.Aizencang GI, Solis C, Bishop DF, Warner C, Desnick RJ. Human uroporphyrinogen III synthase: genomic organization, alternative promoters and erythroid-specific expression. Genomics. 2000;70:223–231. doi: 10.1006/geno.2000.6373. [DOI] [PubMed] [Google Scholar]
- 12.2000. Human gene mutation database. Institute of Medical Genetics. Cardiff, Wales, United Kingdom. http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html.
- 13.Warner CA, Yoo HW, Roberts AG, Desnick RJ. Congenital erythropoietic porphyria: identification and expression of exonic mutations in the uroporphyrinogen III synthase gene. J Clin Invest. 1992;89:693–700. doi: 10.1172/JCI115637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, Warner CA, Desnick RJ. Congenital erythropoietic porphyria: identification and expression of 10 mutations in the uroporphyrinogen III synthase gene. J Clin Invest. 1995;95:905–912. doi: 10.1172/JCI117742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontanellas A, et al. A systematic analysis of the mutations of the uroporphyrinogen III synthase gene in congenital erythropoietic porphyria. Eur J Hum Genet. 1996;4:274–282. doi: 10.1159/000472214. [DOI] [PubMed] [Google Scholar]
- 16.Xu W, Astrin KH, Desnick RJ. Molecular basis of congenital erythropoietic porphyria: mutations in the human uroporphyrinogen III synthase gene. Hum Mutat. 1996;7:187–192. doi: 10.1002/(SICI)1098-1004(1996)7:3<187::AID-HUMU1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Anderson MA, Gusella JF. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984;20:856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.2000. Chromosome 10 genetic map. Center for Medical Genetics. Marshfield, Wisconsin, USA. http://research.marshfieldclinic.org/genetics.
- 20.Gelb BD, Edelson JG, Desnick RJ. Linkage of pycnodysostosis to chromosome 1q21 by homozygosity mapping. Nat Genet. 1995;10:235–237. doi: 10.1038/ng0695-235. [DOI] [PubMed] [Google Scholar]
- 21.Martignetti JA, et al. The May-Hegglin anomaly (MHA) gene localizes to a <1 Mb region on chromosome 22q12.3-13.1. Am J Hum Genet. 2000;66:1449–1454. doi: 10.1086/302873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottgens B, et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 23.Boulechfar S, et al. Heterogeneity of mutations in the uroporphyrinogen III synthase gene in congenital erythropoietic porphyria. Hum Genet. 1992;88:320–324. doi: 10.1007/BF00197267. [DOI] [PubMed] [Google Scholar]
- 24.Lim LC, Fang L, Swendeman SL, Sheffery M. Characterization of the molecularly cloned murine alpha-globin transcription factor CP2. J Biol Chem. 1993;268:18008–18017. [PubMed] [Google Scholar]
- 25.Yoon JB, Li G, Roeder RG. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. doi: 10.1128/mcb.14.3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swendeman SL, et al. Characterization of the genomic structure, chromosomal location, promoter, and development expression of the alpha-globin transcription factor CP2. J Biol Chem. 1994;269:11663–11671. [PubMed] [Google Scholar]
- 27.Tsai SF, et al. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- 30.Tsang AP, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 31.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang N, Miller WL. Cloning of factors related to HIV-inducible LBP proteins that regulate steroidogenic factor-1-independent human placental transcription of the cholesterol side-chain cleavage enzyme, P450scc. J Biol Chem. 2000;275:2852–2858. doi: 10.1074/jbc.275.4.2852. [DOI] [PubMed] [Google Scholar]
- 34.Zambrano N, Minopoli G, de Candia P, Russo T. The Fe65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1. J Biol Chem. 1998;273:20128–20133. doi: 10.1074/jbc.273.32.20128. [DOI] [PubMed] [Google Scholar]
- 35.Ikuta T, Kan YW, Swerdlow PS, Faller DV, Perrine SP. Alterations in protein-DNA interactions in the gamma-globin gene promoter in response to butyrate therapy. Blood. 1998;92:2924–2933. [PubMed] [Google Scholar]
- 36.Jane SM, Nienhuis AW, Cunningham JM. Hemoglobin switching in man and chicken is mediated by a heteromeric complex between the ubiquitous transcription factor CP2 and a developmentally specific protein [erratum 1995, 14:854] EMBO J. 1995;14:97–105. doi: 10.1002/j.1460-2075.1995.tb06979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aizencang GI, Bishop DF, Forrest D, Astrin KH, Desnick RJ. Uroporphyrinogen III synthase: an alternative promoter controls erythroid-specific expression in the murine gene. J Biol Chem. 2000;275:2295–2304. doi: 10.1074/jbc.275.4.2295. [DOI] [PubMed] [Google Scholar]
- 38.Semenza, G.L. 1998. Transcription factors and human disease. Oxford University Press. New York, New York, USA. 368 pp.
- 39.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 40.Koivisto UM, Palvimo JJ, Janne OA, Kontula K. A single-base substitution in the proximal Sp1 site of the human low density lipoprotein receptor promoter as a cause of heterozygous familial hypercholesterolemia. Proc Natl Acad Sci USA. 1994;91:10526–10530. doi: 10.1073/pnas.91.22.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun XM, Neuwirth C, Wade DP, Knight BL, Soutar AK. A mutation (T-45C) in the promoter region of the low-density-lipoprotein (LDL)-receptor is associated with a mild clinical phenotype in a patient with heterozygous familial hypercholesterolaemia (FH) Hum Mol Genet. 1995;4:2125–2129. doi: 10.1093/hmg/4.11.2125. [DOI] [PubMed] [Google Scholar]
- 42.Cooper DN, Krawczak M. The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- 43.Forget BG. Molecular basis of hereditary persistence of fetal hemoglobin. Ann NY Acad Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 44.Reijnen MJ, Sladek FM, Bertina RM, Reitsma PH. Disruption of a binding site for hepatocyte nuclear factor 4 results in hemophilia B Leyden. Proc Natl Acad Sci USA. 1992;89:6300–6303. doi: 10.1073/pnas.89.14.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reijnen MJ, Peerlinck K, Maasdam D, Bertina RM, Reitsma PH. Hemophilia B Leyden: substitution of thymine for guanine at position -21 results in a disruption of a hepatocyte nuclear factor 4 binding site in the factor IX promoter. Blood. 1993;82:151–158. [PubMed] [Google Scholar]
- 46.Picketts DJ, Lillicrap DP, Mueller CR. Synergy between transcription factors DBP and C/EBP compensates for a haemophilia B Leyden factor IX mutation. Nat Genet. 1993;3:175–179. doi: 10.1038/ng0293-175. [DOI] [PubMed] [Google Scholar]
- 47.Picketts DJ, Mueller CR, Lillicrap D. Transcriptional control of the factor IX gene: analysis of five cis-acting elements and the deleterious effects of naturally occurring hemophilia B Leyden mutations. Blood. 1994;84:2992–3000. [PubMed] [Google Scholar]
- 48.2000 Genetic Location Database. Department of Human Genetics, University of Southhampton, Southampton, United Kingdom. http://cedar.genetics.soton.ac.uk/public_html/ldb.html.ß
- 49.2000 CEPH Genotype Database. Jean Dausset Foundation - CEPH, Paris, France. http://landru.cephb.fr/cephdb/.