Many pathogens have evolved the ability to alter surface-exposed molecules, most often in response to selective pressures associated with the host immune system (1). Pathogenic bacteria exhibit numerous examples of this adaptive strategy, and a range of molecular mechanisms has evolved in these bacteria for generating genetic variation at individual loci termed “contingency loci” (2). Many contingency genes are controlled by simple sequence DNA repeats that accumulate reversible, rec-independent mutations at high frequency. A striking feature of the complete genome sequences of the human pathogens Haemophilus influenzae and Neisseria meningitidis is the abundance of loci containing simple sequences (3, 4). Intriguingly, both of these bacteria are obligate commensals of the upper respiratory tract of humans but can, in some hosts, cause life-threatening invasive disease. In this Perspective we provide a synthesis of our current understanding of how polymorphisms in simple sequence contingency loci produce phenotypic variation. We also evaluate the role of this variation in pathogenesis and the evolution of virulent bacterial strains.
Bacterial adaptation, genetic variation, and contingency loci
All organisms face the challenge of maintaining their fitness in diverse and changing environments. The key to fitness resides in the capacity to change biological characteristics. Such phenotypic variation can be achieved by regulating gene expression or through genetic change. While gene regulation offers a powerful mechanism for producing rapid phenotypic change, it is deterministic and does not offer as flexible a solution for providing novel variants as does random genetic change. In the case of pathogenic bacteria, the host environment is a particularly stringent test of their adaptive potential, owing to the polymorphisms and acquired immune mechanisms of their hosts. Two features of the bacterial lifestyle give pathogens an advantage in the production of genetic variation that can contribute positively to the host-microbe “arms race”: the potentially large size of bacterial populations and their rapid rates of replication. The ability to quickly generate large numbers of progeny increases the probability that at least one cell in any population will have a mutation that will allow it to adapt to any new environment. However, in many bacterial infections, including those caused by H. influenzae and N. meningitidis, genetic variation is restricted because infections often result from clonal expansion of a few founder organisms and because population sizes and replication rates are limited by the availability of essential nutrients and by competition for them from other microbes. These restrictions have been overcome by the evolution of molecular mechanisms that rapidly generate large amounts of genetic variation.
Contingency loci and the biology of “microbial anticipation”
“But there are also responses of genomes to unanticipated challenges that are not so precisely programmed. The genome is unprepared for these shocks. Nevertheless, they are sensed and the genome responds in a discernible but initially unforeseen manner.” As Barbara McClintock noted (5), genomes can respond to unanticipated challenges. This is because natural selection has selected genomes that best survived unexpected challenges in the past. For many bacterial species, this flexibility results from the presence in the genome of hypermutable contingency loci, which provide a repertoire of variation, allowing the population to adapt rapidly in the face of unpredictable contingencies, such as changes in the host environment (2). These loci generate large amounts of genetic variation at genes that have a disproportionately large impact on microbial fitness, and this process operates in a stochastic manner. Although this hypermutability can result from several mechanisms (2), there is growing interest in those loci containing tandem DNA repeats (microsatellites) with unit sizes of 1–8 bp. We define these regions as “simple sequence contingency loci.”
These loci contain tandem repeats within either a coding sequence or a promoter; altered numbers of repeats thus allow for switches in the reading frame of translation or changes in the level of promoter activity. Two features of these loci distinguish them from other mechanisms of phenotypic variation. First, the hypermutation of these loci is independent of classic recombination genes, in contrast to other mechanisms of phase variation such as gene conversion, transposition, and “flip-flop.” Second, the reversibility and hypermutability of simple sequences result from expansion or contraction of the repeat tracts by one or more repeat units, such that there are multiple possible “ON” and “OFF” states. For example, the gain or loss of one or two repeat units could switch an ON tetranucleotide repeat tract in a reading frame to OFF. Because these mutations can occur in a small number of generations, these loci can appear highly polymorphic even between cells of a single clone.
The simple sequence contingency loci of H. influenzae and N. meningitidis
The genomes of H. influenzae strain Rd and N. meningitidis strains MC58 and Z2491 have been completely sequenced (6–8). There are many potential simple sequence contingency loci in these genomes, with estimates of 12 for the H. influenzae sequence (9, 10) and 26 to 65 for the Neisseria sequences (3, 8). Some of these loci have yet to be functionally characterized, but many cause phase variation of factors involved in microbe-host interactions, allowing the bacteria to alter their pattern of adherence to or invasion of host cells or their means of scavenging essential nutrients. Phase variation can therefore provide a repertoire of variants from which the fittest bacterial cells are selected. However, the infection process is a dynamic one, and variants that are fit at one stage and in a particular host location may not be so at a later stage or in a different econiche. In parallel with these considerations, host clearance mechanisms, innate and acquired, can target surface molecules. Thus, the fitness of a variant must be measured not merely by its functional contribution but by the extent to which it is targeted by the host’s immune clearance mechanisms.
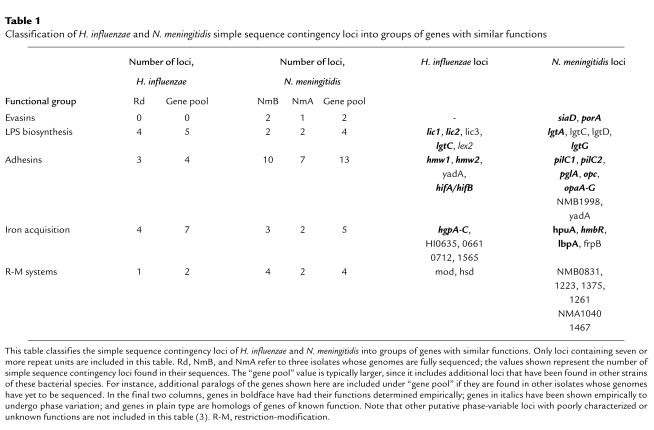
Clearly, phase variation can increase fitness by allowing bacteria to evade the host’s immune system. The function and antigenicity of a given molecule, although seemingly independent properties, are often under coordinate selection in vivo, particularly when the bacterial genome includes multiple loci with redundant functions. In this case, the population can easily generate variants that escape immune responses while maintaining essential functions. Figure 1 shows a hypothetical model of how genomes containing multiple simple sequence contingency loci with similar functions permit a clonal bacterial infection to go through multiple rounds of expansion of different antigenic variants any one of which can carry out a specific function. The number of rounds of expansion will depend on the number of antigenically different genes with overlapping function that are present in a genome. To facilitate discussion of the biological effects of simple sequence contingency loci, we have categorized the loci into five functional groups: evasins, LPS biosynthesis genes, adhesins, iron acquisition proteins, and restriction-modification systems (Table 1).
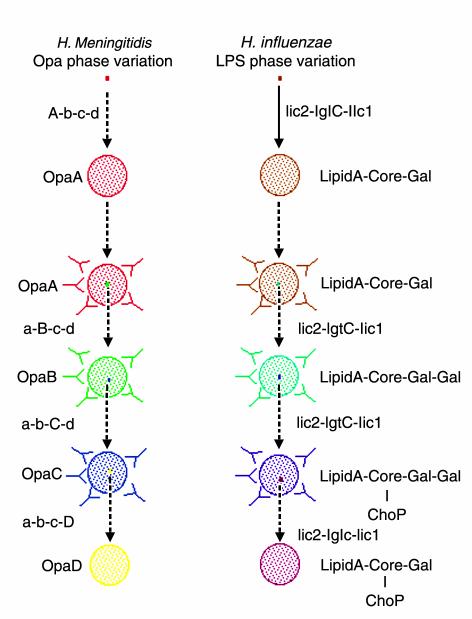
Figure 1.
Generation of antigenic variation by simple sequence contingency loci. Simple sequence contingency loci can rapidly generate variants during infections started by small numbers of bacteria that are essentially clonal. The events shown in (a) and (b) depict the hypothetical progression of two surface features that are targeted by the host immune system. In each, a single bacterium undergoes between 5 and 25 generations (solid line) to form a population within which there are a number of variants (27). After an indeterminate time (dotted lines), the host raises an antibody response against one determinant (a) or one part of a determinant (b) of these bacteria. A variant with an antigenically different determinant then escapes this immune response and replicates to form a new population containing variants that can then escape a second immune response. This process is repeated multiple times. (a) The opa genes of N. meningitidis provide an example of switching between determinants of similar function. Multiple, antigenically distinct forms of bacterial surface adhesins may be expressed in turn during the course of an infection, allowing the bacteria to evade the host’s immune defenses. (b) The LPS biosynthesis genes provide an example of switching between different modifications of a single determinant. The genotypes of the founder cell and the variants are indicated alongside the lines; genes that are switched on at a given point are shown in capital letters. The phenotype of the majority of cells in a population is indicated alongside the circles. In a, the product is a protein (note that the designations A to D for the opa genes [and proteins] are arbitrary and do not indicate particular opa genes); in b, the loci produce modifications to a basal LPS structure (16).
Table 1.
Classification of H. influenzae and N. meningitidis simple sequence contingency loci into groups of genes with similar functions
Phase-variable evasins.
Capsule is the major determinant of invasive disease for N. meningitidis. In vitro, the expression of capsule allows bacteria to resist phagocytosis and killing by complement pathways (11) but also interferes with attachment (12). Phase variation of capsule is controlled, in serogroup B strains, by a mononucleotide repeat tract in the siaD gene, whose product is involved in capsule biosynthesis (13). In meningococcal carriage, noncapsular bacteria compose a significant proportion of the population in the upper respiratory tract, whereas capsular forms predominate in invasive sites (14). In animal models, unencapsulated meningococci undergo phase variation to capsular forms during systemic invasion (15). Thus, capsule phase variation may be adaptive because the absence of capsule in nasopharyngeal bacteria allows for efficient adhesion and the outgrowth of encapsulated forms allows the population to survive despite the presence of host defenses such as opsonophagocytosis and complement. This switch between two different phenotypes may be critical for transmission, establishment, and maintenance of infections by serogroup B strains of N. meningitidis.
Phase-variable LPS biosynthesis proteins.
LPS is the major constituent of the outer surface of Gram-negative bacteria and is intimately involved in every stage of pathogenesis (reviewed in refs. 16, 17). It has multiple roles in attachment of bacterial cells to host membranes and in regulating bacterial resistance to the innate immune system. Phase variation between different LPS phenotypes is critical for pathogenesis. LPS also elicits a strong antibody response from the human host and is therefore expected to be under strong selective pressure to produce antigenic variation. In both H. influenzae and N. meningitidis, enzymes required for the addition of modifications to the exposed surfaces of LPS molecules are encoded by simple sequence contingency loci.
Bacterial genomes with multiple simple sequence contingency loci that differentially modify a single determinant, such as LPS, have a powerful resource for creating antigenic variation. For instance, an H. influenzae strain containing all five phase-variable LPS biosynthesis genes (see Table 1) can potentially produce 32 genotypes. If each of these LPS molecules is antigenically different, this strain could go through 32 rounds of clonal expansion of unique antigenic variants (Figure 1b). However, LPS is a complex molecule that can present several epitopes to the host immune system simultaneously, and not all of the phase-variable modifications produced in both organisms are equally immunogenic (16–18). It is essential, therefore, to determine how each of these potential genotypes effects both the antigenicity and the function of LPS molecules.
In H. influenzae, the lic1 locus, which contains four genes, is required for addition of phosphorylcholine (ChoP) to LPS (18, 19). Phase variation of this locus, as detected by the presence or absence of this modification, is controlled by changes in the length of the repeat tract in lic1A. Presence of ChoP on LPS of some H. influenzae strains confers sensitivity to C-reactive protein–mediated complement killing but resistance to an antimicrobial peptide found in the upper respiratory tract (20). Lysenko et al. (18) have demonstrated selection for ChoP-positive phase variants in a rat model of infection, and indeed, a high percentage of respiratory tract isolates from natural infections have lic1 in frame, suggesting that the presence of ChoP on LPS is required for persistence on mucosal surfaces. Thus, it appears that the lic1 locus mediates phase variation between a phenotype in which colonization is enhanced and one in which immune resistance is increased (18).
N. meningitidis LPS structures can be differentiated using monoclonal antibodies into 12 immunotypes, which correlate, albeit imperfectly, with different LPS structures. Six genes, four of which (lgtA, lgtC, lgtD, and lgtG) are phase-variable, act in different combinations to generate these immunotypes (21), and the presence or absence of these six genes determines the potential repertoire of immunotypes that a strain can produce. The immunotypes produced at a particular time are determined by the ON/OFF status of the phase-variable genes. Phase-variable expression of these genes means that strains can switch immunotypes. Further variation comes from sialylation of LPS, which can occur on only particular immunotypes. This modification inhibits phagocytosis and complement-mediated killing of N. meningitidis but also interferes with Opa/Opc-mediated adhesion to and invasion of host cells (reviewed in ref. 17). Phase variation between immunotypes with and without the potential to be sialylated is correlated with invasive disease in animal models. Consistent with this finding, most carriage isolates in natural infections are of nonsialylated immunotypes, but invasive isolates are of immunotypes with the potential to be sialylated.
Phase-variable adhesins.
Both H. influenzae and N. meningitidis have multiple simple sequence contingency loci with adhesion phenotypes (Table 1), and most of these adhesins elicit antibody responses during natural infections by these bacteria (12, 22, 23). These multiple loci may, if their attachment properties overlap, allow these bacteria to go through multiple rounds of expansion of different antigenic variants, thus escaping host immune responses, without loss of the ability to adhere. Conversely, if the attachment properties differ, phase variation may alter the tissue tropism of variants and permit adaptation to new niches. The Opa proteins of N. meningitidis provide examples of both these scenarios.
N. meningitidis genomes normally contain three or four opa genes, whose products mediate attachment to and invasion of various host cells and tissues (reviewed in ref. 23). Most meningococcal Opa proteins bind to members of the CEACAM receptor family of proteins, with some Opa’s binding to individual receptors and others to multiple receptors (24). Phase variation of these proteins is due to the presence of pentanucleotide repeats within the reading frame of these genes, and variation of Opa protein expression has been observed during nasopharyngeal carriage (25). Although the overlapping attachment properties of Opas permit antigenic variation (see Figure 1a), differing molecular specificities may mean that Opas have different tissue tropisms. Thus, phase variation of these proteins could permit the bacteria to adapt to different niches during the course of an infection.
Phase-variable iron acquisition proteins.
Iron acquisition is critical to bacterial growth, and hence to pathogenesis. The necessity for pathogenic bacteria to scavenge this resource efficiently has resulted in a redundancy of mechanisms, involving numerous surface-expressed proteins. Both H. influenzae and N. meningitidis have multiple simple sequence contingency loci whose products mediate acquisition of iron (Table 1). Phase variation of these loci may produce antigenic variation similar to that proposed for the opa genes (see Figure 1a and above). Alternatively, if the sources of iron vary between body compartments, such that proteins with different specificities are required for each compartment, phase variation may allow adaptation to new niches (26).
Phase-variable restriction-modification systems.
Contingency loci encoding homologs of restriction-modification systems form a unique category unrelated to those described so far. Only type I and type III restriction enzymes appear to be good candidates for phase-variable gene expression (3, 9, 27), probably because these systems only function as complexes of subunits. The biological role usually ascribed to restriction enzymes is defense against infection by bacteriophages. It is unclear whether bacteriophages exert a strong selective pressure on respiratory pathogens, but having multiple restriction systems with different combinations of systems switched on in each cell may be one mechanism whereby bacteria resist attack by bacteriophages (28).
Allelic variation and simple sequence contingency loci
Above we discussed the consequences of switching of simple sequence contingency loci, and we noted that genomes containing multiple loci can, through switching, generate large amounts of genetic and phenotypic diversity. We now wish to consider the influence of allelic variation, or interstrain polymorphisms in natural populations of H. influenzae and N. meningitidis, on the generation of diversity by these loci. In this discussion, it is important to distinguish between polymorphisms that affect the number of phenotypes generated by simple sequence contingency loci and those that affect the rate at which this diversity is generated.
Influence of allelic variations in simple sequence contingency loci on phenotypic variation.
Strains exhibit polymorphism in the numbers and combinations of simple sequence contingency genes present in their genomes (3, 21). Variation also occurs through the horizontal transfer between strains of whole loci, as is seen in the capsule switching that can occur during meningococcal epidemics (29). Many polymorphisms are known to alter the antigenicity of phase-variable proteins in H. influenzae (22) and in N. meningitidis (30). For example, Opa proteins undergo microevolution within clonal populations of meningococcal epidemics (30). Some polymorphisms in simple sequence contingency loci alter the function of the product of a locus. In H. influenzae, there are two alleles of lic1D that differ in the position on the LPS molecule to which ChoP is attached; these LPS molecules then differ in the amount of C-reactive protein that binds to them (18). These two alleles, therefore, generate different antigenic and functional forms of LPS.
Influence of allelic variations on the rate of phase variation of simple sequence contingency loci.
Mutation rates of microsatellites or simple sequences are controlled by the repeat tracts and surrounding sequences and by the products of other genes (i.e., trans-acting factors). Many polymorphisms have been found in the repeat tracts of simple sequence contingency loci in natural populations of H. influenzae and N. meningitidis. For example, a survey of H. influenzae clinical isolates found wide variations in repeat numbers for the LPS biosynthesis genes (3–32 for lic1; 12–36 for lic2; 7–36 for lgtC; and 12–36 for lic3) and other loci (9). In N. meningitidis, the number of pentanucleotide repeats in opa genes varies from 7 to 18 copies (30) and the number of mononucleotides in the poly(G) tract of lgtA varies from 5 to 14 (21). Phase variation rates have been measured for a number of simple sequence contingency loci using antibodies or reporter constructs. Although these studies provide important characterizations of individual loci, they do not indicate the effects of polymorphisms in repeat tract length on phase variation rate. Recently, De Bolle et al. (27) performed a more extensive analysis of the relationship between repeat tract length and phase variation rate, using a reporter gene in the mod locus, and found that rates increased proportionately with tract length over the range 17 to 38 repeat units (27). Thus, the observed polymorphisms between strains in repeat tract lengths can alter mutation rates, raising the possibility that the rate of phase variation for individual simple sequence contingency loci is itself under selection.
In principle, polymorphisms in genes encoding trans-acting factors may affect the mutation rates of one or multiple loci. However, only one such polymorphism has been described. Capsule phase variation rates vary between N. meningitidis strains. Bucci et al. (31) found that strains with high phase variation rates had identical deletions in the dam gene, whose product is a DNA methylase. These authors showed that deletion of dam from strains with an intact allele increased capsule phase variation rate. This dam mutation was present in all pathogenic strains but in only 50% of commensal strains for one collection of isolates, suggesting a link between hypermutation and virulence (31). More recent studies, however, have not confirmed the overrepresentation of the dam mutation among pathogenic strains of Neisseria (32–34).
Implications of allelic variations in mutation rates of simple sequence contingency loci.
De Bolle et al. (27) have developed a theoretical model that simulates switching of three phase-variable loci to examine the effects of altering the rates of phase variation at simple sequence contingency loci. They find that a threefold increase in mutation rate produces a ninefold increase in the genetic diversity of bacterial populations over time scales commensurate with that of an infection in humans. Thus, the observed variations in lengths of repeat tracts and polymorphisms in genes encoding trans-acting factors can have dramatic effects on the rate at which variants are produced in infections. Survival may ultimately depend upon the rate at which rare variants can be generated in the bacterial population (35).
Mutator strains and the evolution of virulence
H. influenzae and N. meningitidis are respiratory pathogens/commensals whose founder and carriage populations are likely to be restricted in size. Transmission depends upon aerosol or intimate exchange of fomites and — compared with fecal-oral spread of enteric bacteria, for example — is likely to be inefficient and to result in population bottlenecks. These small founder inocula have to survive rapidly fluctuating environments and host clearance mechanisms, placing a high premium on the evolution of mechanisms like simple sequence contingency locus variation that can rapidly introduce stochastic genetic and phenotypic changes (35). The amount of variation is amplified by the presence of multiple loci of similar function within individual genomes and by polymorphisms in these loci between strains. Polymorphisms in trans-acting factors are particularly interesting, since the dam mutation, the only described example in these species, also increases global mutation rates. Such “mutator” strains, which have been studied in other bacterial species, can profoundly affect bacterial adaptation and the evolution of virulence (36, 37). Mutators allow for synergistic effects on the levels of phenotypic variation, because they increase both phase variation rates of simple sequence contingency loci and global mutation rates.
Simple sequence contingency loci often encode host-interactive products or virulence factors. The large amount of phenotypic variation produced by these loci is likely, therefore, to contribute to the maintenance of bacterial fitness and hence to the evolution of virulence. In particular, the presence of multiple simple sequence contingency loci within each genome and within the gene pool means that these species have a vast number of genotypes with which to explore “host landscapes” in the struggle to survive in a hostile environment. Genotypes that survive best may include those with a large complement of virulence-associated phase-variable genes and high mutation rates. These genotypes may have an increased chance of generating variants that have a greater potential to cause disease. Thus, the rate of generation of variants and the types of variants generated may be major factors in determining the risk that a particular strain will cause disease.
Acknowledgments
This work was supported by a grant from the Wellcome Trust. D. Field is a Sloan–National Science Foundation postdoctoral Fellow in Molecular Evolution.
References
- 1.Brunham RC, Plummer FA, Stephens RS. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect Immun. 1993;61:2273–2276. doi: 10.1128/iai.61.6.2273-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moxon ER, Rainey PB, Nowak MA, Lenski R. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 3.Saunders NJ, et al. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 4.Hood DW, et al. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 6.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 7.Tettelin H, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 8.Parkhill J, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–505. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 9.van Belkum A, et al. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect Immun. 1997;65:5017–5027. doi: 10.1128/iai.65.12.5017-5027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood DW, et al. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]
- 11.Vogel U, Frosch M. Mechanisms of neisserial serum resistance. Mol Microbiol. 1999;32:1133–1139. doi: 10.1046/j.1365-2958.1999.01469.x. [DOI] [PubMed] [Google Scholar]
- 12.Nassif X, Pujol C, Morand P, Eugene E. Interactions of pathogenic Neisseria with host cells. Is it possible to assemble the puzzle? Mol Microbiol. 1999;32:1124–1132. doi: 10.1046/j.1365-2958.1999.01416.x. [DOI] [PubMed] [Google Scholar]
- 13.Hammerschmidt S, et al. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialytransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 14.Cartwright, K. 1995. Meningococcal carriage and disease. In Meningococcal disease. K. Cartwright, editor. John Wiley and Sons. Chichester, United Kingdom. 115–146.
- 15.Mackinnon FG, Gorringe AR, Funnell SG, Robinson A. Intranasal infection of infant mice with Neisseria meningitidis. Microb Pathog. 1992;12:415–420. doi: 10.1016/0882-4010(92)90004-8. [DOI] [PubMed] [Google Scholar]
- 16.Hood, D., and Moxon, E.R. 1999. Lipopolysaccharide phase variation in Haemophilus and Neisseria. In Endotoxin in health and disease. H. Brude, S.M. Opal, S.N. Vogel, and D.C. Morrison, editors. Marcel Dekker Inc. New York, New York, USA. 39–54.
- 17.van Putten JPM, Robertson BD. Molecular mechanisms and implications for infection of lipopolysaccharide variation in Neisseria. Mol Microbiol. 1995;16:847–853. doi: 10.1111/j.1365-2958.1995.tb02312.x. [DOI] [PubMed] [Google Scholar]
- 18.Lysenko E, et al. The position of phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae affects binding and sensitivity to C-reactive protein mediated killing. Mol Microbiol. 2000;35:234–245. doi: 10.1046/j.1365-2958.2000.01707.x. [DOI] [PubMed] [Google Scholar]
- 19.Weiser JN, Shchepetov M, Chong STH. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect Immun. 1997;65:943–950. doi: 10.1128/iai.65.3.943-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lysenko ES, Gould J, Bals R, Wilson JM, Weiser JN. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect Immun. 2000;68:1664–1671. doi: 10.1128/iai.68.3.1664-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jennings MP, et al. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology. 1999;145:3013–3021. doi: 10.1099/00221287-145-11-3013. [DOI] [PubMed] [Google Scholar]
- 22.Gilsdorf JR. Antigenic diversity and gene polymorphisms in Haemophilus influenzae. Infect Immun. 1998;66:5053–5059. doi: 10.1128/iai.66.11.5053-5059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehio C, Gray-Owen SD, Meyer TF. The role of neisserial Opa proteins in interactions with host cells. Trends Microbiol. 1998;6:489–495. doi: 10.1016/s0966-842x(98)01365-1. [DOI] [PubMed] [Google Scholar]
- 24.Muenzer P, et al. Carcinoembryonic antigen family receptor specificity of Neisseria meningitidis Opa variants influences adherence to and invasion of proinflammatory cytokine-activated endothelial cells. Infect Immun. 2000;68:3601–3607. doi: 10.1128/iai.68.6.3601-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods JP, Cannon JG. Variation in expression of class 1 and class 5 outer membrane proteins during nasopharyngeal carriage of Neisseria meningitidis. Infect Immun. 1990;58:569–572. doi: 10.1128/iai.58.2.569-572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren Z, Jin H, Whitby PW, Morton DJ, Stull TL. Role of CCAA nucleotide repeats in regulation of hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J Bacteriol. 1999;181:5865–5870. doi: 10.1128/jb.181.18.5865-5870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Bolle X, et al. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol Microbiol. 2000;35:211–222. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 28.Raleigh EA. Restricting parasites: why are there so many restriction endonucleases? The NEB Transcript (scientific newsletter from New England Biolabs) 1998;9:6–7. [Google Scholar]
- 29.Swartley JS, et al. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobbs MM, Seiler A, Achtman M, Cannon JG. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol Microbiol. 1994;12:171–180. doi: 10.1111/j.1365-2958.1994.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 31.Bucci C, et al. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol Cell. 1999;3:435–445. doi: 10.1016/s1097-2765(00)80471-2. [DOI] [PubMed] [Google Scholar]
- 32.Sun, L., et al. 2000. Many, but not all, invasive isolates of Neisseria meningitidis (NM) have insertional inactivation of dam by drg.Abstract Guide for the 12th International Pathogenic Neisseria Conference. Galveston, Texas, USA. p. 53.
- 33.Klee, S., Kusecek, B., Wintermeyer, E., and Achtman, M. 2000. Mismatch repair in Neisseria meningitidis: higher mutation and phase variation frequencies due to the absence of the dam methyltransferase? Abstract Guide for the 12th International Pathogenic Neisseria Conference. Galveston, Texas, USA. p. 90.
- 34.Cannon, J.G., et al. 2000. Does DNA mismatch repair affect the virulence of pathogenic Neisseria species? Abstract Guide for the 12th International Pathogenic Neisseria Conference. Galveston, Texas, USA. p. 86.
- 35.Bayliss CD, Field D, De Bolle X, Moxon ER. The generation of diversity by Haemophilus influenzae. Response. Trends Microbiol. 2000;8:435–436. doi: 10.1016/s0966-842x(00)01841-2. [DOI] [PubMed] [Google Scholar]
- 36.Moxon ER, Thaler DS. The tinkerer’s evolving tool-box. Nature. 1997;387:659–662. doi: 10.1038/42607. [DOI] [PubMed] [Google Scholar]
- 37.Radman M. Enzymes of evolutionary change. Nature. 1999;401:866–868. doi: 10.1038/44738. [DOI] [PubMed] [Google Scholar]