Abstract
Type IA and type II DNA topoisomerases are distinguished by their ability to cleave one or two strands, respectively, of a DNA duplex. Both types have been proposed to use an “enzyme-bridging” mechanism, in which a break is formed in a DNA strand and a gap is opened between the broken pieces to allow passage of a second DNA strand or duplex segment. Although the type IA and type II topoisomerase structures appear overall quite different from one another, unexpected similarities between several structural elements suggest that members of the two subfamilies may use comparable mechanisms to bind and cleave DNA.
Keywords: DNA cleavage/DNA binding/catabolite-activator protein-like domain/Rossmann fold
Topoisomerases are enzymes that can alter DNA topology. Their activity requires cleavage and religation of DNA. The cleavage step generates a covalent phosphotyrosine link between an enzyme and a DNA strand; the religation step restores the phosphodiester bond. Topoisomerases have been classified into two primary types, I and II, according to their ability to cleave one or both strands, respectively, of a DNA duplex (1). Type I enzymes are further subdivided into two classes, IA and IB, based on differences in their amino acid sequences and reaction mechanisms (for a recent review, see ref. 2).
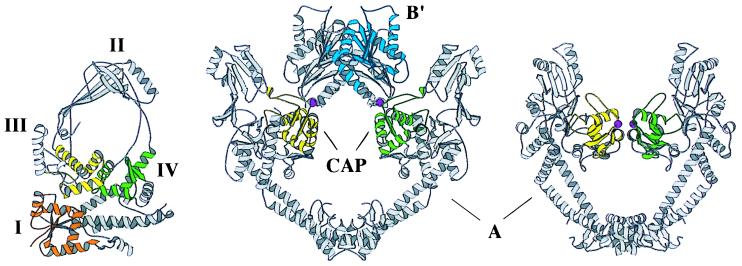
The three-dimensional structures of the DNA-binding/cleavage cores of Escherichia coli topoisomerase I (a type IA protein) and two type II proteins, Saccharomyces cerevisiae topoisomerase II and E. coli GyrA, have been determined by X-ray crystallography (Fig. 1) (3–5). In addition to the binding/cleavage core, type II enzymes also have an ATPase function that is required for full catalytic activity, and the structure of the ATP-binding domain of E. coli GyrB has been determined (6). As expected from amino acid sequence, the overall tertiary structures of the type IA and type II enzymes appear to be very different. The DNA-binding/cleavage region of type IA enzymes is monomeric, with a large globular base capped by a tall arch (Fig. 1 Left), whereas in the type II enzymes, this region is dimeric, having two crescent-shaped halves that meet to encompass a large hole (Fig. 1 Center and Right).
Figure 1.
Comparison of E. coli DNA topoisomerase I (Left), S. cerevisiae DNA topoisomerase II (Middle), and E. coli GyrA (Right). Regions discussed in the text are highlighted in color: CAP-like part of DNA topoisomerase I domain III (yellow), CAP-like part of DNA topoisomerase I domain IV (green), DNA topoisomerase II and GyrA CAP-like domains (yellow and green), Rossmann fold of DNA topoisomerase I domain I (orange), Rossmann fold of DNA topoisomerase II B′ domain (blue), and active-site tyrosines of all topoisomerases (magenta spheres). Figure generated by molscript (49).
Despite their structural and functional differences, however, type IA and type II topoisomerases display some noteworthy mechanistic similarities. Enzymes of both types form 5′ phosphotyrosine links to DNA. Both alter DNA superhelicity in discrete steps per round of DNA cleavage and rejoining (the change in linking number being 1 or 2, respectively, for type IA or type II enzymes) (7, 8). By contrast, type IB enzymes form 3′ phosphotyrosine links to DNA and appear to change linking number by variable integral amounts per cleavage/rejoining event (9). Moreover, both type IA and type II, but not type IB, enzymes require divalent metal ions (preferably Mg2+) for full activity (8, 10–14).
A particularly important similarity between the type IA and type II enzymes is their capacity to catenate or decatenate DNA rings (8, 15). This activity probably reflects their role as “untangling” enzymes in the cell, to enable chromosomal segregation. It requires that the enzyme create and bridge a break in a DNA segment, while subsequently allowing a second DNA segment to pass between the two broken ends. Both ends of the broken DNA remain associated with the enzyme by the covalent 5′ phosphotyrosine link and by noncovalent binding of the 3′-hydroxyl end. Therefore, the type IA and type II topoisomerases have been proposed to serve as “enzyme bridges” that span transient single-stranded or double-stranded DNA breaks, respectively (8, 16).
This paper reports the results of a detailed comparative inspection of regions that are thought to interact with the target DNA in type IA and type II enzymes. We have identified local structural similarities between the two groups that may explain some of their functional similarities. In particular, several subdomains known to be important for DNA cleavage show a degree of structural and chemical relatedness generally found between protein homologs of similar function. The results raise the possibility that, in terms of DNA binding, cleavage, and bridging, type IA and type II enzymes may share a common catalytic mechanism.
MATERIALS AND METHODS
The program o, version 6.2 (17), was used to obtain a preliminary visual superposition of structural elements and to select polypeptide regions for comparison. Root-mean-squared deviations (rmsd) of atom positions for E. coli DNA topoisomerase I (BNL Protein Data Bank accession no. 1ecl), S. cerevisiae DNA topoisomerase II (Brookhaven National Laboratory Protein Data Bank accession no. 1bgw), and E. coli GyrA (coordinates kindly provided by A. Maxwell and R. Liddington, Leicester University) were calculated by using the program lsqkab from the CCP4 suite (18, 19). Tables 1a and 2a outline the regions compared with one another. As a blind check, domain coordinates were submitted to the DALI server (20) for comparison against a known structural fold database. Z-scores from DALI are probabilistic indicators of the quality of fit, with scores of Z > 2.0 implying structural similarity.
Table 1.
Comparison of topoisomerase CAP-like domains
| a. | Residue range, CAP-like domains*
|
||||
|---|---|---|---|---|---|
| α1 | α2 | α3 | β2 | β3 | |
| Topo I DIII | 385–398† | 286–297 | 300–311 | 315–320 | 367–371 |
| Topo I DIV | 195–208 | 479–490 | 493–504 | 508–513 | 516–520 |
| Topo II | 702–715 | 722–733 | 738–749 | 760–765 | 785–789 |
| GyrA | 43–56 | 66–77 | 81–92 | 101–106 | 124–128 |
| b. | Results of structural comparison
|
|
|---|---|---|
| CAP-like domain rmsd [from lsqkab (18)] | DALI Z-score | |
| Topo I (DIII v. DIV) | 2.0 | 7.3‡ |
| GyrA vs. Topo I DIII | 1.7 | 4.2 |
| GyrA vs. Topo I DIV | 2.2 | 3.5§ |
| GyrA vs. Topo II | 1.0 | 11.1 |
Because of the level of divergence between topoisomerases, β1 was omitted from these comparisons.
Note that despite being located C-terminal to the other elements of the fold, this helix in domain III occupies the same spatial position as the first helix in other CAP-like folds.
DALI matched domain III to domain IV only after renumbering domain III such that the last helix of the domain becomes the first. This renumbering does not swap the N- to C-terminal polarity of the helix.
Using the GyrA CAP domain as a search object in DALI obtained a match for domain IV but not for domain III of DNA topoisomerase I.
Table 2.
Comparison of topoisomerase α/β domains
| a | Residue range, Rossmann-like folds
|
|||||||
|---|---|---|---|---|---|---|---|---|
| β1 | α1 | β2 | α2 | β3 | α3 | β4 | α4 | |
| Topo I | 2–10 | 11–20 | 23–31 | 87–101 | 103–111 | 119–128 | 133–139 | 146–152 |
| Topo II | 443–451 | 452–461 | 468–476 | 490–504 | 519–527 | 535–544 | 556–562 | 613–619 |
| b | Results of structural comparison
|
|
|---|---|---|
| α-β domain rmsd [from lsqkab (18)] | DALI Z-score | |
| Topo I vs. Topo II | 2.3 | 7.0 |
RESULTS AND DISCUSSION
There are three published structures of type IA and type II DNA topoisomerase fragments that can bind DNA and that contain the active-site tyrosine: E. coli DNA topoisomerase I, E. coli GyrA subunit, and S. cerevisiae DNA topoisomerase II (4, 5, 21). Comparative structural overviews are shown in Fig. 1.
The type IA enzymes are generally single-chain, monomeric proteins; the E. coli DNA topoisomerase I structure of this class includes residues 2 to 590 of the 865 amino acid protein. This fragment can cleave single-stranded DNA, but it lacks a C-terminal region that aids DNA binding and is important for full catalytic activity. There are four distinct domains in DNA topoisomerase I (2–590): an α/β “Rossmann-fold” (domain I) (22), a small β-barrel (domain II), and two domains (III and IV) with folds similar to one found in the E. coli catabolite-activator protein (CAP) (23, 24) (Fig. 1). The active-site tyrosine responsible for DNA cleavage is in domain III. This tyrosine is buried in the structure, suggesting that the protein is in a conformational state similar but not identical to the one in which DNA is bound but not cleaved and opened (21).
In prokaryotes, type II enzymes are A2B2 tetramers. The B subunit contains the ATPase activity, whereas the A subunit contains the DNA-cleaving, active-site tyrosine. In eukaryotic type II topoisomerases, the A and B subunits are part of a single polypeptide chain, and the enzymes are (B-A)2 dimers, with the B region N-terminal to the A region. The yeast DNA topoisomerase II and E. coli GyrA structures (Fig. 1) comprise most of the A regions of the enzymes, including residues 678–1177 of the yeast enzyme and residues 30–522 of GyrA. These binding and cleavage cores are termed the A′ fragments. The yeast DNA topoisomerase II structure also contains nearly one-half of the B region, residues 419–628, termed B′. The conformations of the A′ regions in the yeast DNA topoisomerase II and GyrA structures are different, such that the spatial relationship of the active-site tyrosines is changed. The yeast DNA topoisomerase II structure may represent a state similar to the one in which a DNA duplex has been cleaved and the two halves separated, whereas the GyrA structure appears to be a state that can bind uncleaved duplex DNA (4, 5).
Spatial Relationship of CAP-Like Domains that Bear the Active-Site Tyrosines.
The DNA-binding domain of the E. coli catabolite-activator protein (CAP) is a common structural scaffold found in a large number of DNA-binding proteins (for reviews see refs. 25–27). This domain generally contains a three α-helix bundle backed by a three- or four-stranded β-sheet. The order of secondary structural elements is usually αβααββ, but also can be βααββα, in which the last helix retains the spatial position and chain polarity of what is generally the first helix. The two adjacent a-helices (usually the second and third helices of the bundle) are connected by a short turn and are oriented with respect to each other such that their helical axes cross at an angle of ≈120°. This motif is known as the helix–turn–helix (HTH), and it is responsible for many of the principal contacts between CAP-like proteins and their DNA targets. The second helix of this motif generally inserts into the major groove of DNA, and the turn contacts the phosphodiester backbone.
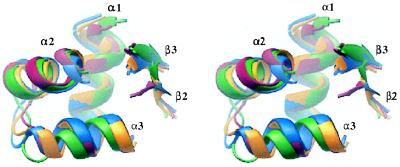
It has been observed previously that domains III and IV of E. coli DNA topoisomerase I and the A′ fragments of DNA topoisomerase II and GyrA all contain α-β regions with CAP-like folds (4, 24). The active-site tyrosines of these enzymes are always on an extended loop that connects the adjacent β-strands. This loop is in an analogous position to the “wing” of the “winged-helix” motif, a substructure that is found in certain CAP-like domains such as HNF-3 (28) and that contacts the DNA phosphodiester backbone. Domain IV of DNA topoisomerase I and the CAP-like domains of DNA topoisomerase II and GyrA have the same αβααββ order as CAP, but domain III of DNA topoisomerase I is βααββα. Moreover, in domain III, although the adjacent β-strands are too short to meet the Kabsch and Sander secondary structure criteria (29), the central HTH motif is nonetheless very clear. Table 1 shows the results of superpositions of segments of the CAP-like domains from DNA topoisomerase I, DNA topoisomerase II, and GyrA. As expected, the type II CAP-like folds superpose on each other quite well (Table 1, Fig. 2), but they also superpose relatively well on domains III and IV of DNA topoisomerase I (Fig. 2), as well as on other CAP-like members such as histone H5 (not shown) (30).
Figure 2.
Stereo view of the superposed backbones of CAP-like regions from E. coli DNA topoisomerase I domain III (red), DNA topoisomerase I domain IV (green), S. cerevisiae DNA topoisomerase II (blue), and E. coli GyrA (yellow). The domains are oriented such the second helix of the HTH motif is facing forward. Figure generated by ribbons (50).
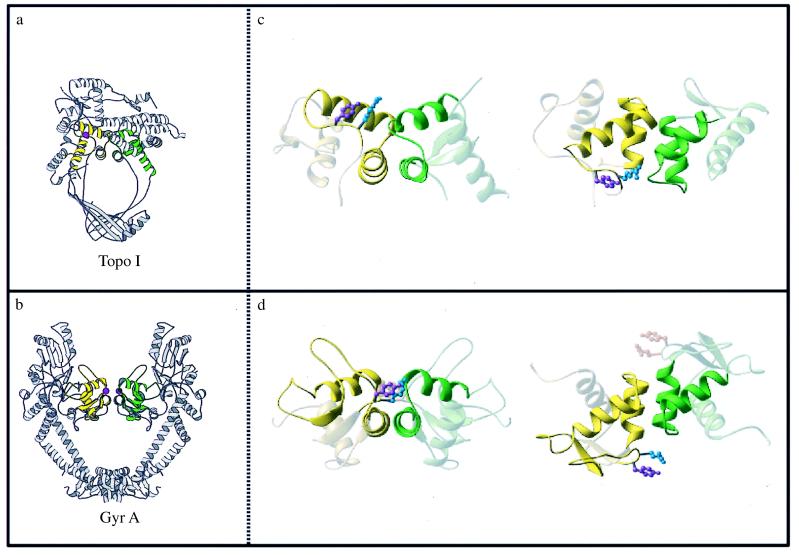
A striking feature of the DNA topoisomerase I and GyrA structures, not previously noticed, is a similar spatial relationship of the CAP-like domains. The HTH motifs of domains III and IV of DNA topoisomerase I are related by a pseudo twofold axis; in GyrA they are related by a true dyad (Fig. 3). In both cases, the first helix of each HTH motif (α2) packs antiparallel to its twofold-related partner. This arrangement places the active-site tyrosine close to the turn of the HTH of the other CAP-like domain. In DNA topoisomerase I, there is only one such tyrosine, whereas in GyrA, there is a symmetrically related pair.
Figure 3.
Arrangement of the CAP-like domains found in type Ia and type II proteins. (a and b) Representations of the DNA topoisomerase I (a) and GyrA (b) structures. Domains III and IV of DNA topoisomerase I are colored yellow and green, respectively. The CAP-like domains of GyrA also are colored yellow and green. The active-site tyrosines of the molecules are represented as magenta spheres. (c and d) Close-up views of the CAP-like domains of DNA topoisomerase I (c) and GyrA (d). The HTH motifs of the CAP-like domains are highlighted for clarity, and the active-site tyrosine and a conserved, neighboring arginine are drawn in ball-and-stick representations. The left-hand images in c and d are viewed exactly as in a and b, respectively; the right-hand images in c and d are “top” views, looking down the (pseudo) twofold axis. (a and b) Generated by molscript (49); (c and d) made by ribbons (50).
The two tyrosines in the published GyrA structure are too far apart to bridge comfortably the width of a DNA duplex, and Liddington and coworkers (5) have proposed that this type II enzyme distorts the DNA when it binds. The type IA enzymes are known to bind junctions of duplex and single-stranded DNA, as well as single-stranded segments (31, 32). The similar orientations of the CAP-like domains in DNA topoisomerase I and GyrA place the two recognition helices adjacent to one another, and accommodating both helices in the major groove, may require the DNA to unwind locally as an initial step in the catalytic mechanism.
The similar orientation of the type IA and type II CAP-like domains also may explain their common DNA cleavage polarity. Both type IA and type II enzymes generate 5′ phosphotyrosine linkages to DNA, and it is likely that the active-site tyrosines attack the DNA backbone from a similar local geometry. Type IA and type II enzymes also have an absolutely conserved arginine next to the active-site tyrosine (Arg-321 in E. coli DNA topoisomerase I, Arg-781 in yeast DNA topoisomerase II, and Arg-121 in GyrA), suggesting that this residue may have a role in DNA binding or cleavage. In support of this idea, mutation of Arg-321 of the E. coli DNA topoisomerase I or of Arg-781 of yeast DNA topoisomerase II to alanine results in reduced DNA relaxation and cleavage activity (ref. 33 and Q. Liu and J.C.W., unpublished results). Thus, the common use of twofold or pseudo twofold-related CAP domains and certain conserved residues for DNA binding and cleavage by type IA and type II enzymes may reflect a fundamentally similar organization of the catalytic complex.
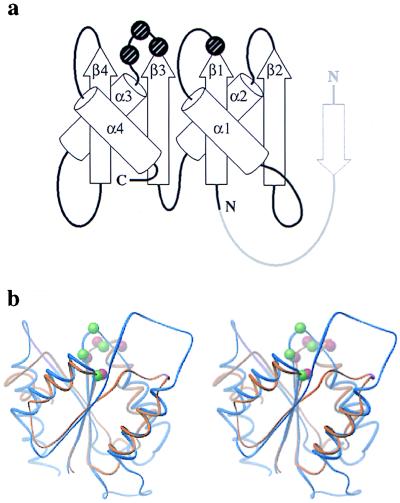
Domain I of DNA Topoisomerase I and the B′ Region of DNA Topoisomerase II Are Based on a Similar Fold.
The central lobe of the B′ domain of DNA topoisomerase II and domain I of DNA topoisomerase I are both α-β structures, with a four-stranded, parallel β-sheet sandwiched between two pairs of α-helices. In both proteins, the βαβαβαβα fold has the same order and relative positioning of secondary structural elements (Fig. 4a); this fold resembles the “Rossmann fold” found in a number of proteins known to bind nucleotides, phosphorylated amino acids, or other small organic molecules (22). In DNA topoisomerase I, this domain contacts domains III and IV directly and lies just to one side of the active-site tyrosine. The proximity of domain I to the active-site tyrosine and its topological similarity to known nucleotide-binding domains have led to the proposal that it assists in binding the 3′-OH end of the DNA after cleavage by the tyrosine (21). In type II enzymes, the B′ region is known to be important for DNA binding and cleavage (4, 34, 35). Certain conserved amino acid sequence motifs within this region of DNA topoisomerase II have been predicted to constitute a nucleotide-binding motif (36), whereas in bacterial DNA gyrase, residues within these sequence motifs can be mutated to confer resistance to quinolones that stabilize the covalent DNA gyrase/DNA complex (37).
Figure 4.
Similarities of the type II B′ core and the type IA domain I. (a) Folding diagrams of DNA topoisomerase I domain I and the DNA topoisomerase II B′ domain. The position of the acidic quartet of amino acids is marked by spheres. The DNA topoisomerase II B′ domain has an additional small β-strand (drawn in light gray) that lies at the edge of the overall fold. (b) Wire stereo diagram [ribbons (50)] showing the superposition of the two domains; DNA topoisomerase I is orange and DNA topoisomerase II blue. The Cα positions of the acidic residues are shown as red (DNA topoisomerase I) and green (DNA topoisomerase II) spheres.
One sequence comparison had previously suggested that portions of the B′ region of type II enzymes were related to the domain I region of DNA topoisomerase I (38). The superposed structures show, however, that the N-terminal regions of the sequences were misaligned (Fig. 4b). The misalignment in the earlier sequence analysis probably arose from the presence of large insertions (60–200 amino acids) between the secondary-structural elements of the folds of both type IA and type II enzymes. In support of the correct structure-based realignment between the type-IA domain I and the type-II B′ fragment (Table 2), submission of the DNA topoisomerase I domain I coordinates to DALI (a structural comparison and alignment database) (20), returns the DNA topoisomerase II B′ domain as the closest structural analog, with a probability score well above other nucleotide binding motifs (Z = 7.0, compared with a score of Z = 5.3 for the next best fit, p21-Ras, and Z = 4.5 for Che Y).
There is also a chemical similarity between domain I of topoisomerase I and the topoisomerase II B′ domain. Both domains contain a nearly invariant set of acidic residues having identical spacing and occupying the same position within the domains. The acidic sequences lie within 4–8 Å of each other and include an E/D-X-E/D-X-E/D pattern in the β3-α3 loop and a conserved glutamic acid at the β1/α1 junction (Fig. 4b). In E. coli DNA topoisomerase I, these residues (Glu-9, Asp-111, Asp-113, and Glu-115) lie close to the active-site tyrosine, and it has been proposed that they coordinate Mg2+ in an analogous manner to DNA polymerases (21, 39). In DNA topoisomerase II, these acidic residues (Glu-449, Asp-526, Asp-528, and Asp-530) are part of several highly conserved signature sequence motifs for type II enzymes. Mutation of Glu-9 in domain I of DNA topoisomerase I or Glu-449 and Asp-530 in DNA topoisomerase II severely impairs DNA breakage and rejoining activity (ref. 33 and Q. Liu and J.C.W., unpublished results).
Structural similarities between the type IA and II Rossmann folds and the Che Y protein support the notion of an acidic metal-binding site. Che Y, which binds phospho-histidine and coordinates transfer of the phosphate to an acceptor aspartate residue (40, 41), has a set of acidic residues (Asp-12, Asp-13, and Asp-57), which coordinate Mg2+ to assist in catalysis (42). These residues lie at the β1/α1 and β3/α3 junctions of the α/β fold, like the acidic residues in the topoisomerases. The “open” nature of the metal coordinating center of the CheY fold allows divalent metal ions such as Mn2+ to substitute for the preferred Mg2+ ion (43, 44). Type IA and type II topoisomerases can likewise use other divalent metals for DNA cleavage and even for limited catalysis (12, 13, 45), suggesting that topoisomerases also may possess an open metal binding site coordinated by key acidic residues.
The structural and chemical similarities between domain I of DNA topoisomerase I and the B′ region of type II DNA topoisomerases suggest related functions during catalysis, but the orientation of the B′ domain in the published yeast DNA topoisomerase II structure projects the acidic residues away from the active-site tyrosine and the proposed site of DNA-binding, rather than toward it, as in DNA topoisomerase I (Fig. 1). There is ample evidence that type II enzymes pass through distinct structural states during catalysis (46–48). Could the relative position of domains I, III, and IV in topoisomerase I suggest an alternate orientation for the topoisomerase B′ domain? To examine this possibility, we superposed the CAP-like domains of topoisomerase IA on GyrA. The superposition allowed the subsequent positioning of the type II B′ domains based on the location of topoisomerase I domain I (Fig. 5). This speculative, structure-based reorganization places the conserved acidic groups from the type II B′ domain in the neighborhood of the active-site tyrosine. We note that the acidic cluster is located close to two known quinolone-resistance “hot-spots” in E. coli DNA GyrB, which have been proposed to lie near the active-site tyrosine (37). A new structure of topoisomerase II recently has been determined, which shows that the B′ domains can indeed undergo large-scale reorientations with respect to the A′ fragments (D.F. and J.M.B., unpublished data).
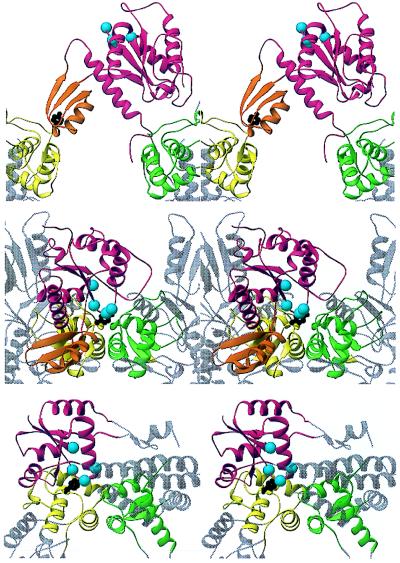
Figure 5.
Comparison of a hypothetical hybrid A′/B′ model (Middle) with the yeast DNA topoisomerase II structure (Top) and DNA topoisomerase I (Bottom). The hybrid type II model was generated by combining the A′ conformation observed in the GyrA structure (5) with a B′ orientation based on that seen for domain I (relative to Tyr-319) in DNA topoisomerase I (21). Note that this new orientation can be produced by a simple rotation of the B′ domain and that the tether connecting B′ to A′ would permit such a motion. The B′ core and domain I are colored red. The α/β extension of the B′core is orange. The Cα positions of the conserved acidic cluster are marked by cyan-colored spheres. The CAP-like domains are colored yellow and green, and one active-site tyrosine is shown in black. Figure generated using ribbons (50).
CONCLUSION
Our structural comparison has uncovered some previously unsuspected similarities between type IA and type II DNA topoisomerases. The published structures of E. coli DNA topoisomerase I and GyrA are thought to represent states that bind to uncleaved DNA, and this work shows that the two CAP-like domains (domains III and IV) of DNA topoisomerase I are so oriented that they resemble the dyad-related CAP-like domains in GyrA. Moreover, the position of the single active-site tyrosine of DNA topoisomerase I with respect to these CAP-like folds is close to the position of the analogous active-site tyrosines of GyrA. The central lobe of the B′ domain of yeast DNA topoisomerase II closely resembles domain I of DNA topoisomerase I, not only in its α/β fold but also in the placement of conserved acidic residues that have been suggested to bind Mg2+ for activity in DNA topoisomerase I. The two groups of enzymes may thus share certain properties of their DNA binding, cleavage, and bridging activities through the use of common structural motifs displayed on distinct supporting elements. An appreciation of the structural and mechanistic similarities between these two distinct topoisomerase families should foster a more comprehensive understanding of type IA and type II topoisomerase function.
Acknowledgments
We thank J. Lindsley for careful reading of drafts of the manuscript, R. Liddington and A. Maxwell for coordinates of GyrA, and S. Lynch and P. S. Kim for discussion. J.M.B. acknowledges support from the W. M. Keck Foundation and the Whitehead Institute; D.F., support from National Institutes of Health Grant PO1 HL41484-10 (to P. Kim); J.C.W., support from National Institutes of Health Grant GM24544. S.C.H. is an Investigator with the Howard Hughes Medical Institute.
ABBREVIATIONS
- CAP
catabolite-activator protein
- HTH
helix–turn–helix
References
- 1.Liu L F, Liu C C, Alberts B M. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 2.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 3.Lima C D, Wang J C, Mondragón A. Nature (London) 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 4.Berger J M, Gamblin S J, Harrison S C, Wang J C. Nature (London) 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 5.Cabral J H M, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Nature (London) 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 6.Wigley D B, Davies G J, Dodson E J, Maxwell A, Dodson G. Nature (London) 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown P O, Cozzarelli N R. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 8.Brown P O, Cozzarelli N R. Proc Natl Acad Sci USA. 1981;78:843–847. doi: 10.1073/pnas.78.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stivers J T, Harris T K, Mildvan A S. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 10.Gellert M, Mizuuchi K, O’Dea M H, Nash H A. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champoux J J. Annu Rev Biochem. 1978;47:449–479. doi: 10.1146/annurev.bi.47.070178.002313. [DOI] [PubMed] [Google Scholar]
- 12.Goto T, Laipis P, Wang J C. J Biol Chem. 1984;16:10422–10429. [PubMed] [Google Scholar]
- 13.Osheroff N. Biochemistry. 1987;26:6402–6406. doi: 10.1021/bi00394a015. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C-X, Roche C J, Tse-Dihn Y-C. J Biol Chem. 1997;272:16206–16210. doi: 10.1074/jbc.272.26.16206. [DOI] [PubMed] [Google Scholar]
- 15.Mizuuchi K, Fisher L M, O’Dea M H, Gellert M. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J C. In: Nucleases. Roberts R, Linn S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 41–57. [Google Scholar]
- 17.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch W. Acta Crystallogr A. 1976;32:922–923. [Google Scholar]
- 19.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 20.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 21.Lima C D, Wang J C, Mondragón A. J Mol Biol. 1993;232:1213–1216. doi: 10.1006/jmbi.1993.1474. [DOI] [PubMed] [Google Scholar]
- 22.Branden C, Tooze J. Introduction to Protein Structure. New York: Garland; 1991. [Google Scholar]
- 23.Schultz S C, Shields G C, Steitz T A. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 24.Murzin A G. Curr Opin Struct Biol. 1994;4:441–449. [Google Scholar]
- 25.Harrison S C. Nature (London) 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 26.Pabo C, Sauer R T. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 27.Nelson H C M. Curr Opin Genet Dev. 1995;5:180–189. doi: 10.1016/0959-437x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 28.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 30.Ramakrishnan V, Finch J T, Graziano V, Lee P L, Sweet R M. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 31.Dean F B, Krasnow M A, Otter R, Matzuk M M, Spengler S J, Cozzarelli N R. Cold Spring Harbor Symp Quant Biol. 1983;47:769–777. doi: 10.1101/sqb.1983.047.01.088. [DOI] [PubMed] [Google Scholar]
- 32.Kirkegaard K, Pflugfelder G, Wang J C. Cold Spring Harbor Symp Quan Biol. 1984;49:411–419. doi: 10.1101/sqb.1984.049.01.047. [DOI] [PubMed] [Google Scholar]
- 33.Chen S-J, Wang J C. J Biol Chem. 1998;273:6050–6056. doi: 10.1074/jbc.273.11.6050. [DOI] [PubMed] [Google Scholar]
- 34.Brown P O, Peebles C L, Cozzarelli N R. Proc Natl Acad Sci USA. 1979;76:6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gellert M, Fisher L M, O’Dea M H. Proc Natl Acad Sci USA. 1979;83:7152–7156. [Google Scholar]
- 36.Corbett A H, Osheroff N. Chem Res Toxicol. 1993;6:585–597. doi: 10.1021/tx00035a001. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caron P, Wang J C. In: Advances in Pharmacology. Liu L F, editor. New York: Academic; 1994. pp. 271–291. [Google Scholar]
- 39.Beese L S, Steitz T A. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wylie D, Stock A, Wong C-Y, Stock J. Biochem Biophys Res Commun. 1988;151:891–896. doi: 10.1016/s0006-291x(88)80365-6. [DOI] [PubMed] [Google Scholar]
- 41.Hess J F, Oosawa K, Kaplan N, Simon M I. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 42.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 43.Needham J V, Chen T Y, Falke J J. Biochemistry. 1993;32:3363–3367. doi: 10.1021/bi00064a020. [DOI] [PubMed] [Google Scholar]
- 44.Falke J J, Drake S K, Hazard A L, Peersen O B. Q Rev Biophys. 1994;27:219–290. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- 45.Domanico P L, Tse-Dinh Y-C. J Inorg Biochem. 1991;42:87–96. doi: 10.1016/0162-0134(91)80035-g. [DOI] [PubMed] [Google Scholar]
- 46.Osheroff N. J Biol Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 47.Lindsley J E, Wang J C. Proc Natl Acad Sci USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roca J R, Wang J C. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 49.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 50.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]