Abstract
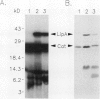
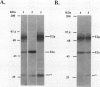
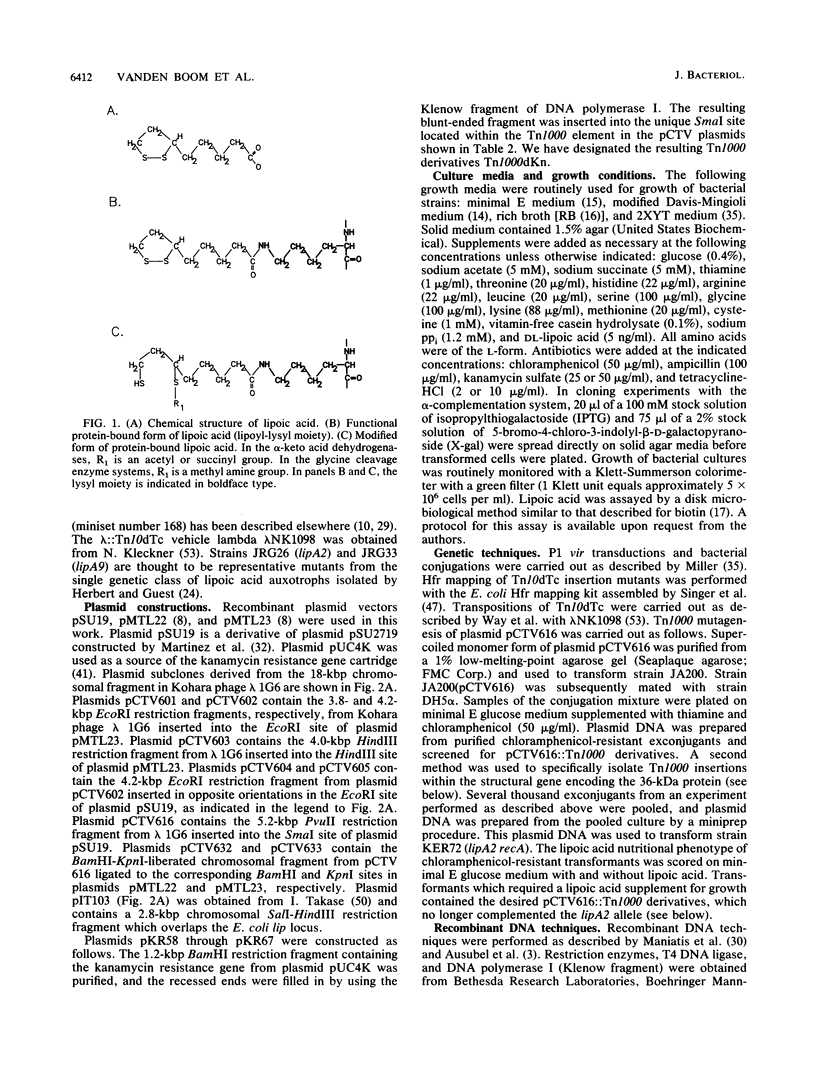
We report the isolation and genetic characterization of novel Tn10dTc and Tn1000dKn insertion mutations in and near the lip locus of the Escherichia coli chromosome. The Tn10dTc and Tn1000dKn mutations define two genes, lipA and lipB, involved in lipoic acid biosynthesis. Two representative alleles (lip-2 and lip-9) from the previously reported genetic class of lipoic acid auxotrophic mutants (A. A. Herbert and J. R. Guest, J. Gen. Microbiol. 53:363-381, 1968) were assigned to the lipA complementation group. We have cloned the E. coli lip locus and developed a recombinant plasmid-based genetic system for fine-structure physical-genetic mapping of mutations in this region of the E. coli chromosome. We also report that a recombinant plasmid containing a 5.2-kbp PvuII restriction fragment from the E. coli lip locus produced three proteins of approximately 8, 12, and 36 kDa by using either a maxicell or in vitro transcription translation expression system. The 36-kDa protein was identified as the gene product encoded by the lipA locus. Finally, we have identified a previously unreported lipoylated protein that functions in the glycine cleavage system of E. coli.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ali S. T., Guest J. R. Isolation and characterization of lipoylated and unlipoylated domains of the E2p subunit of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1990 Oct 1;271(1):139–145. doi: 10.1042/bj2710139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S. T., Moir A. J., Ashton P. R., Engel P. C., Guest J. R. Octanoylation of the lipoyl domains of the pyruvate dehydrogenase complex in a lipoyl-deficient strain of Escherichia coli. Mol Microbiol. 1990 Jun;4(6):943–950. doi: 10.1111/j.1365-2958.1990.tb00667.x. [DOI] [PubMed] [Google Scholar]
- Browner M. F., Taroni F., Sztul E., Rosenberg L. E. Sequence analysis, biogenesis, and mitochondrial import of the alpha-subunit of rat liver propionyl-CoA carboxylase. J Biol Chem. 1989 Jul 25;264(21):12680–12685. [PubMed] [Google Scholar]
- Buck D., Spencer M. E., Guest J. R. Cloning and expression of the succinyl-CoA synthetase genes of Escherichia coli K12. J Gen Microbiol. 1986 Jun;132(6):1753–1762. doi: 10.1099/00221287-132-6-1753. [DOI] [PubMed] [Google Scholar]
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Cronan J. E., Jr, Li S. J., Reed K., Vanden Boom T., Wang A. Y. Locations of the lip, poxB, and ilvBN genes on the physical map of Escherichia coli. J Bacteriol. 1991 Sep;173(17):5258–5259. doi: 10.1128/jb.173.17.5258-5259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. Y., Cronan J. E., Jr Selection for the transfer of phenotypically nonselectable chromosomal mutations to recombinant plasmids through introduction of an altered restriction site. Gene. 1985;40(2-3):353–357. doi: 10.1016/0378-1119(85)90061-7. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J., Porteous C. E. Pyruvate dehydrogenase multienzyme complex of Escherichia coli: determination of the Mr of the lipoate acetyltransferase component. FEBS Lett. 1981 Oct 12;133(1):112–114. doi: 10.1016/0014-5793(81)80483-8. [DOI] [PubMed] [Google Scholar]
- Davis T. N., Muller E. D., Cronan J. E., Jr The virion of the lipid-containing bacteriophage PR4. Virology. 1982 Jul 30;120(2):287–306. doi: 10.1016/0042-6822(82)90031-9. [DOI] [PubMed] [Google Scholar]
- Eckert B., Beck C. F. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J Bacteriol. 1989 Jun;171(6):3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K., Okamura-Ikeda K., Motokawa Y. Chicken liver H-protein, a component of the glycine cleavage system. Amino acid sequence and identification of the N epsilon-lipoyllysine residue. J Biol Chem. 1986 Jul 5;261(19):8836–8841. [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and alpha-ketoglutarate dehydrogenase-less mutants. J Gen Microbiol. 1968 Oct;53(3):363–381. doi: 10.1099/00221287-53-3-363. [DOI] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Lipoic acid content of Escherichia coli and other microorganisms. Arch Microbiol. 1975 Dec 31;106(3):259–266. doi: 10.1007/BF00446532. [DOI] [PubMed] [Google Scholar]
- ITZHAKI R. F., GILL D. M. A MICRO-BIURET METHOD FOR ESTIMATING PROTEINS. Anal Biochem. 1964 Dec;9:401–410. doi: 10.1016/0003-2697(64)90200-3. [DOI] [PubMed] [Google Scholar]
- Kochi H., Kikuchi G. Mechanism of reversible glycine cleavage reaction in Arthrobacter globiformis. Function of lipoic acid in the cleavage and synthesis of blycine. Arch Biochem Biophys. 1976 Mar;173(1):71–81. doi: 10.1016/0003-9861(76)90236-8. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Markiewicz Z., Broome-Smith J. K., Schwarz U., Spratt B. G. Spherical E. coli due to elevated levels of D-alanine carboxypeptidase. Nature. 1982 Jun 24;297(5868):702–704. doi: 10.1038/297702a0. [DOI] [PubMed] [Google Scholar]
- Martinez E., Bartolomé B., de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988 Aug 15;68(1):159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- Meedel T. H., Pizer L. I. Regulation of one-carbon biosynthesis and utilization in Escherichia coli. J Bacteriol. 1974 Jun;118(3):905–910. doi: 10.1128/jb.118.3.905-910.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motokawa Y., Kikuchi G. Glycine metabolism by rat liver mitochondria. IV. Isolation and characterization of hydrogen carrier protein, an essential factor for glycine metabolism. Arch Biochem Biophys. 1969 Dec;135(1):402–409. doi: 10.1016/0003-9861(69)90556-6. [DOI] [PubMed] [Google Scholar]
- Médigue C., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1000 kbp): update I. Mol Microbiol. 1990 Sep;4(9):1443–1454. [PubMed] [Google Scholar]
- Oden K. L., DeVeaux L. C., Vibat C. R., Cronan J. E., Jr, Gennis R. B. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene. 1990 Nov 30;96(1):29–36. doi: 10.1016/0378-1119(90)90337-q. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Thomas J. O. The subunit molecular weights of the alpha-ketoacid dehydrogenase multienzyme complexes from E. coli. FEBS Lett. 1971 Jun 2;15(1):8–12. doi: 10.1016/0014-5793(71)80066-2. [DOI] [PubMed] [Google Scholar]
- Plamann M. D., Rapp W. D., Stauffer G. V. Escherichia coli K12 mutants defective in the glycine cleavage enzyme system. Mol Gen Genet. 1983;192(1-2):15–20. doi: 10.1007/BF00327641. [DOI] [PubMed] [Google Scholar]
- Reed L. J., Hackert M. L. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990 Jun 5;265(16):8971–8974. [PubMed] [Google Scholar]
- Robinson J. R., Klein S. M., Sagers R. D. Glycine metabolism. Lipoic acid as the prosthetic group in the electron transfer protein P2 from Peptococcus glycinophilus. J Biol Chem. 1973 Aug 10;248(15):5319–5323. [PubMed] [Google Scholar]
- SAGERS R. D., GUNSALUS I. C. Intermediatry metabolism of Diplococcus glycinophilus. I. Glycine cleavage and one-carbon interconversions. J Bacteriol. 1961 Apr;81:541–549. doi: 10.1128/jb.81.4.541-549.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G., Boyd A., Stoker N. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J Bacteriol. 1980 Aug;143(2):569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiert P. S., Stauffer L. T., Stauffer G. V. The lpd gene product functions as the L protein in the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1990 Oct;172(10):6142–6144. doi: 10.1128/jb.172.10.6142-6144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase I., Ishino F., Wachi M., Kamata H., Doi M., Asoh S., Matsuzawa H., Ohta T., Matsuhashi M. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987 Dec;169(12):5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Boom T., Cronan J. E., Jr A physical-genetic map of the lipid-containing bacteriophage PR4. Virology. 1990 Jul;177(1):23–32. doi: 10.1016/0042-6822(90)90456-2. [DOI] [PubMed] [Google Scholar]
- Vanden Boom T., Cronan J. E., Jr Genetics and regulation of bacterial lipid metabolism. Annu Rev Microbiol. 1989;43:317–343. doi: 10.1146/annurev.mi.43.100189.001533. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- de Veaux L. C., Clevenson D. S., Bradbeer C., Kadner R. J. Identification of the btuCED polypeptides and evidence for their role in vitamin B12 transport in Escherichia coli. J Bacteriol. 1986 Sep;167(3):920–927. doi: 10.1128/jb.167.3.920-927.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo-Campbell A., Dykhuizen D., Cleary P. P. Enzymic reduction of d-biotin d-sulfoxide to d-biotin. Methods Enzymol. 1979;62:379–385. doi: 10.1016/0076-6879(79)62244-9. [DOI] [PubMed] [Google Scholar]