Abstract
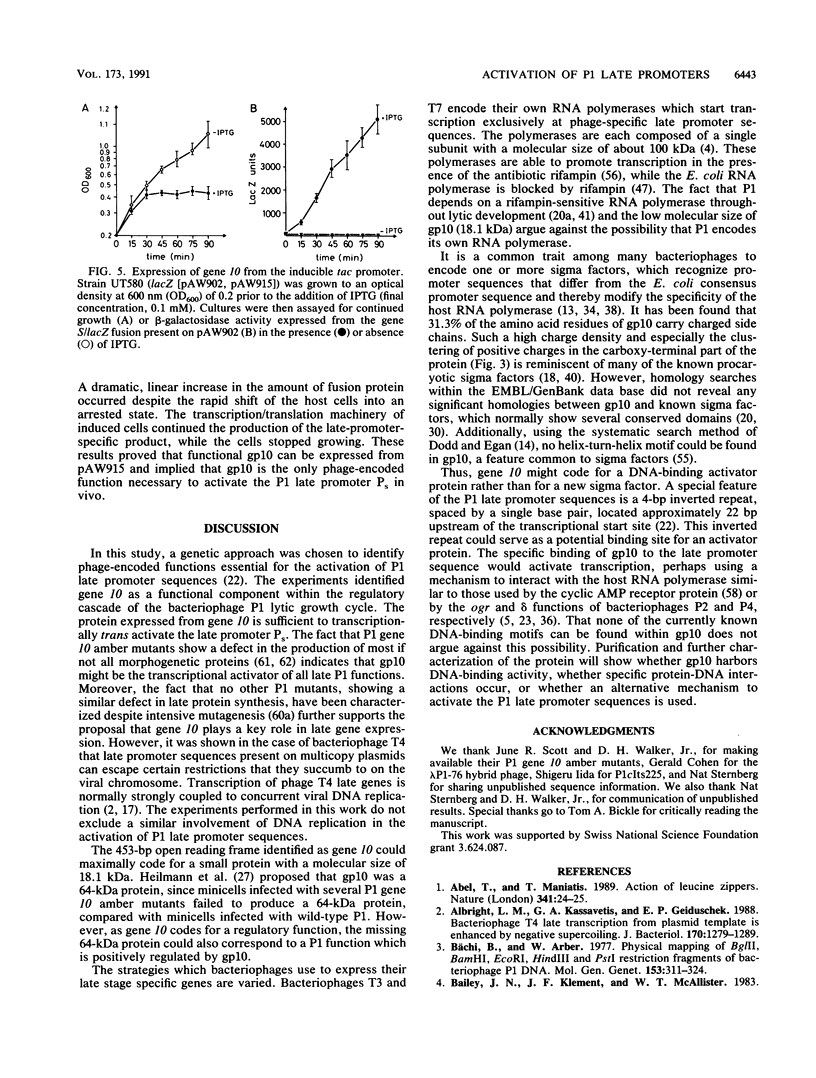
Amber mutants of bacteriophage P1 were used to identify functions involved in late regulation of the P1 lytic growth cycle. A single function has been genetically identified to be involved in activation of the phage-specific late promoter sequence Ps. In vivo, P1 gene 10 amber mutants fail to trans activate a lacZ operon fusion under the transcriptional control of promoter Ps. Several P1 segments, mapping around position 95 on the P1 chromosome, were cloned into multicopy plasmid vectors. Some of the cloned DNA segments had a deleterious effect on host cells unless they were propagated in a P1 lysogenic background. By deletion and sequence analysis, the harmful effect could be delimited to a 869-bp P1 fragment, containing a 453-bp open reading frame. This open reading frame was shown to be gene 10 by sequencing the amber mutation am10.1 and by marker rescue experiments with a number of other gene 10 amber mutants. Gene 10 codes for an 18.1-kDa protein showing an unusually high density of charged amino acid residues. No significant homology to sequences present in the EMBL/GenBank data base was found, and the protein contained none of the currently known DNA-binding motifs. An in vivo trans activation assay system, consisting of gene 10 under the transcriptional control of an inducible promoter and a gene S/lacZ fusion transcribed from Ps, was used to show that gene 10 is the only phage-encoded function required for late promoter activation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abel T., Maniatis T. Gene regulation. Action of leucine zippers. Nature. 1989 Sep 7;341(6237):24–25. doi: 10.1038/341024a0. [DOI] [PubMed] [Google Scholar]
- Albright L. M., Kassavetis G. A., Geiduschek E. P. Bacteriophage T4 late transcription from plasmid templates is enhanced by negative supercoiling. J Bacteriol. 1988 Mar;170(3):1279–1289. doi: 10.1128/jb.170.3.1279-1289.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. N., Klement J. F., McAllister W. T. Relationship between promoter structure and template specificities exhibited by the bacteriophage T3 and T7 RNA polymerases. Proc Natl Acad Sci U S A. 1983 May;80(10):2814–2818. doi: 10.1073/pnas.80.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bächi B., Arber W. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol Gen Genet. 1977 Jun 24;153(3):311–324. doi: 10.1007/BF00431596. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M., Velleman M., Schuster H. Three additional operators, Op21, Op68, and Op88, of bacteriophage P1. Evidence for control of the P1 dam methylase by Op68. J Biol Chem. 1989 Feb 25;264(6):3611–3617. [PubMed] [Google Scholar]
- Cohen G., Sternberg N. Genetic analysis of the lytic replicon of bacteriophage P1. I. Isolation and partial characterization. J Mol Biol. 1989 May 5;207(1):99–109. doi: 10.1016/0022-2836(89)90443-9. [DOI] [PubMed] [Google Scholar]
- D'Ari R., Jaffé-Brachet A., Touati-Schwartz D., Yarmolinsky M. B. A dnaB analog specified by bacteriophage P1. J Mol Biol. 1975 May 25;94(3):341–366. doi: 10.1016/0022-2836(75)90207-7. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Doi R. H., Wang L. F. Multiple procaryotic ribonucleic acid polymerase sigma factors. Microbiol Rev. 1986 Sep;50(3):227–243. doi: 10.1128/mr.50.3.227-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreiseikelmann B., Velleman M., Schuster H. The c1 repressor of bacteriophage P1. Isolation and characterization of the repressor protein. J Biol Chem. 1988 Jan 25;263(3):1391–1397. [PubMed] [Google Scholar]
- Eliason J. L., Sternberg N. Characterization of the binding sites of c1 repressor of bacteriophage P1. Evidence for multiple asymmetric sites. J Mol Biol. 1987 Nov 20;198(2):281–293. doi: 10.1016/0022-2836(87)90313-5. [DOI] [PubMed] [Google Scholar]
- Gitt M. A., Wang L. F., Doi R. H. A strong sequence homology exists between the major RNA polymerase sigma factors of Bacillus subtilis and Escherichia coli. J Biol Chem. 1985 Jun 25;260(12):7178–7185. [PubMed] [Google Scholar]
- Gottesman S., Halpern E., Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981 Oct;148(1):265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
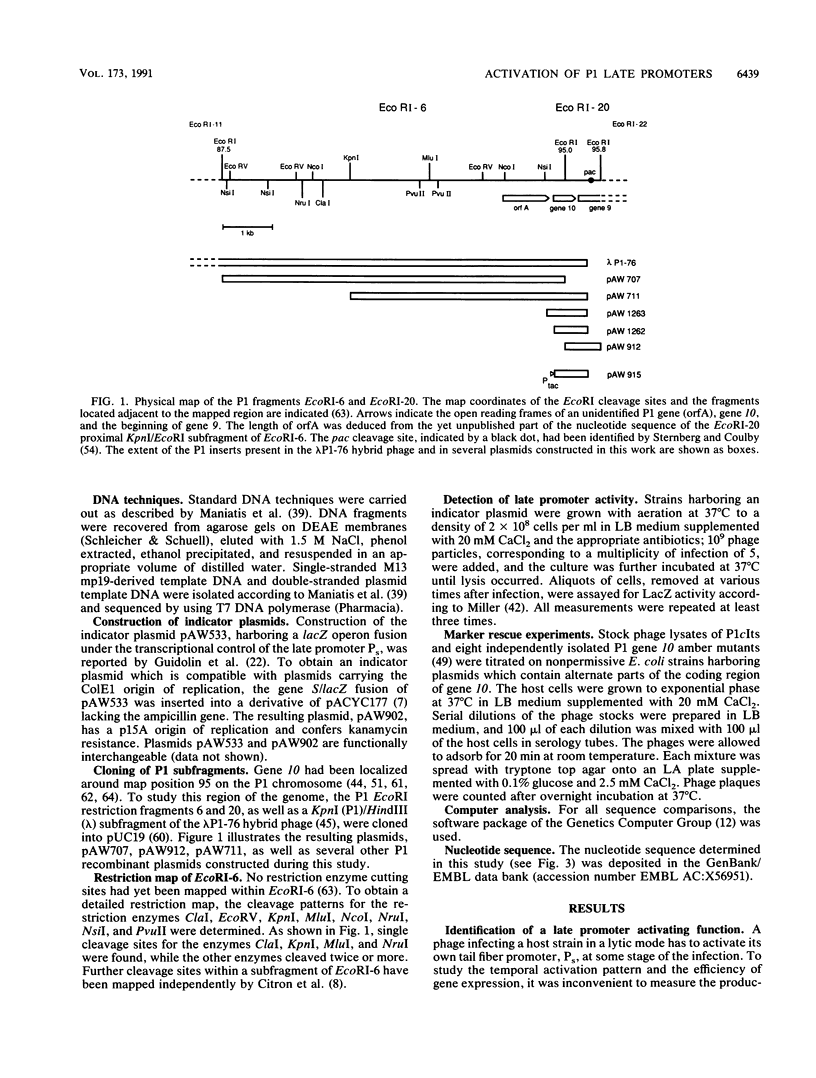
- Guidolin A., Zingg J. M., Arber W. Organization of the bacteriophage P1 tail-fibre operon. Gene. 1989;76(2):239–243. doi: 10.1016/0378-1119(89)90164-9. [DOI] [PubMed] [Google Scholar]
- Guidolin A., Zingg J. M., Lehnherr H., Arber W. Bacteriophage P1 tail-fibre and dar operons are expressed from homologous phage-specific late promoter sequences. J Mol Biol. 1989 Aug 20;208(4):615–622. doi: 10.1016/0022-2836(89)90152-6. [DOI] [PubMed] [Google Scholar]
- Halling C., Calendar R. Bacteriophage P2 ogr and P4 delta genes act independently and are essential for P4 multiplication. J Bacteriol. 1990 Jul;172(7):3549–3558. doi: 10.1128/jb.172.7.3549-3558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiner M., Kempe T., Tjian R. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 1985 Feb 11;13(3):1015–1025. doi: 10.1093/nar/13.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E. B. Structure and regulation of the lytic replicon of phage P1. J Mol Biol. 1989 May 5;207(1):135–149. doi: 10.1016/0022-2836(89)90445-2. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann H., Reeve J. N., Pühler A. Identification of the repressor and repressor bypass (antirepressor) polypeptides of bacteriophage P1 synthesized in infected minicells. Mol Gen Genet. 1980 Apr;178(1):149–154. doi: 10.1007/BF00267223. [DOI] [PubMed] [Google Scholar]
- Heinrich J., Riedel H. D., Baumstark B. R., Kimura M., Schuster H. The c1 repressor of bacteriophage P1 operator-repressor interaction of wild-type and mutant repressor proteins. Nucleic Acids Res. 1989 Oct 11;17(19):7681–7692. doi: 10.1093/nar/17.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T., Velleman M., Schuster H. ban operon of bacteriophage P1. Mutational analysis of the c1 repressor-controlled operator. J Mol Biol. 1989 Jan 5;205(1):127–135. doi: 10.1016/0022-2836(89)90370-7. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Hübner P., Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989 Feb;8(2):577–585. doi: 10.1002/j.1460-2075.1989.tb03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. I. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965 Nov;14(1):85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Zentner P. G., Geiduschek E. P. Transcription at bacteriophage T4 variant late promoters. An application of a newly devised promoter-mapping method involving RNA chain retraction. J Biol Chem. 1986 Oct 25;261(30):14256–14265. [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lee T. C., Christie G. E. Purification and properties of the bacteriophage P2 ogr gene product. A prokaryotic zinc-binding transcriptional activator. J Biol Chem. 1990 May 5;265(13):7472–7477. [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Margolin W., Howe M. M. Localization and DNA sequence analysis of the C gene of bacteriophage Mu, the positive regulator of Mu late transcription. Nucleic Acids Res. 1986 Jun 25;14(12):4881–4897. doi: 10.1093/nar/14.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural R. J., Chesney R. H., Vapnek D., Kropf M. M., Scott J. R. Isolation and characterization of cloned fragments of bacteriophage P1 DNA. Virology. 1979 Mar;93(2):387–397. doi: 10.1016/0042-6822(79)90243-5. [DOI] [PubMed] [Google Scholar]
- Myers D. E., Landy A. The role of host RNA polymerase in P1 phage development. Virology. 1973 Feb;51(2):521–524. doi: 10.1016/0042-6822(73)90456-x. [DOI] [PubMed] [Google Scholar]
- O'Regan G. T., Sternberg N. L., Cohen G. Construction of an ordered overlapping library of bacteriophage P1 DNA in phage vector lambda D69. Gene. 1987;60(1):129–135. doi: 10.1016/0378-1119(87)90220-4. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Riva S., Silvestri L. G. Rifamycins: a general view. Annu Rev Microbiol. 1972;26:199–224. doi: 10.1146/annurev.mi.26.100172.001215. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R. Genetic studies on bacteriophage P1. Virology. 1968 Dec;36(4):564–574. doi: 10.1016/0042-6822(68)90188-8. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Immunity and repression in bacteriophages P1 and P7. Curr Top Microbiol Immunol. 1980;90:49–65. doi: 10.1007/978-3-642-67717-5_3. [DOI] [PubMed] [Google Scholar]
- Sternberg N. A characterization of bacteriophage P1 DNA fragments cloned in a lambda vector. Virology. 1979 Jul 15;96(1):129–142. doi: 10.1016/0042-6822(79)90179-x. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site (pac). I. Differential processing of the cleaved ends in vivo. J Mol Biol. 1987 Apr 5;194(3):453–468. doi: 10.1016/0022-2836(87)90674-7. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site (pac). II. Functional limits of pac and location of pac cleavage termini. J Mol Biol. 1987 Apr 5;194(3):469–479. doi: 10.1016/0022-2836(87)90675-9. [DOI] [PubMed] [Google Scholar]
- Stragier P., Parsot C., Bouvier J. Two functional domains conserved in major and alternate bacterial sigma factors. FEBS Lett. 1985 Jul 22;187(1):11–15. doi: 10.1016/0014-5793(85)81203-5. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
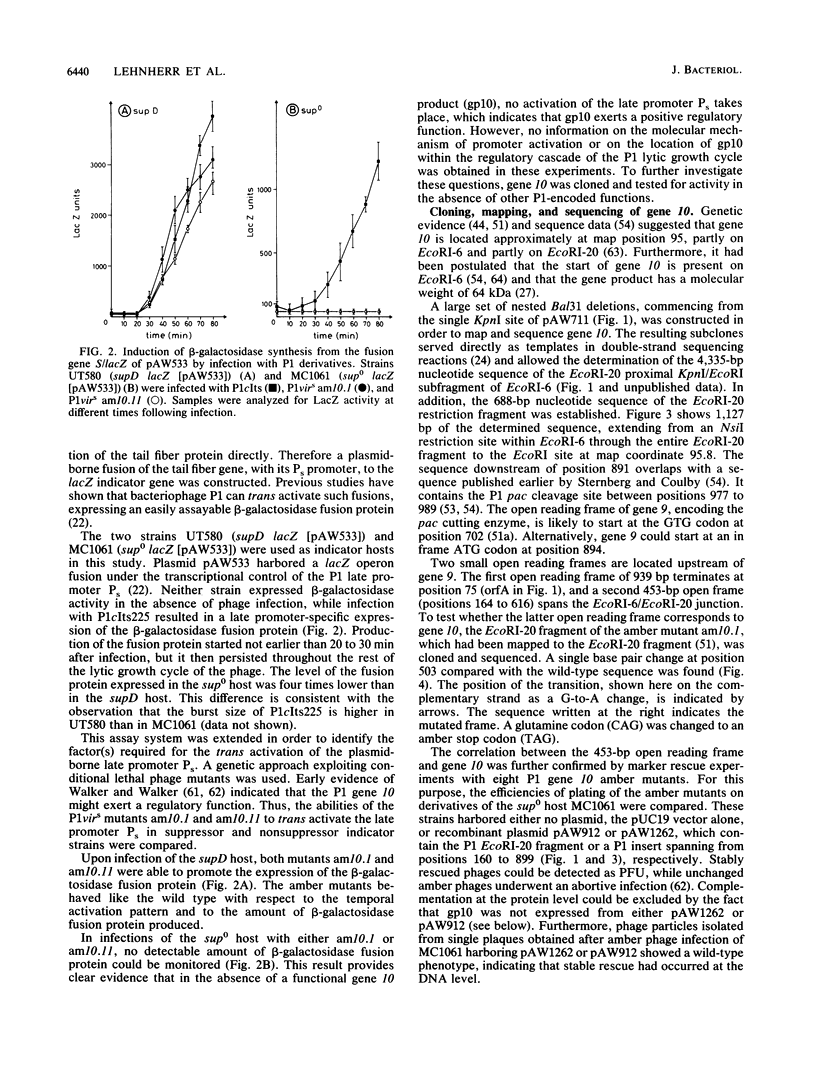
- Susskind M. M., Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978 Jun;42(2):385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velleman M., Dreiseikelmann B., Schuster H. Multiple repressor binding sites in the genome of bacteriophage P1. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5570–5574. doi: 10.1073/pnas.84.16.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walker J. T., Walker D. H., Jr Coliphage P1 morphogenesis: analysis of mutants by electron microscopy. J Virol. 1983 Mar;45(3):1118–1139. doi: 10.1128/jvi.45.3.1118-1139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. T., Walker D. H. Mutations in coliphage p1 affecting host cell lysis. J Virol. 1980 Aug;35(2):519–530. doi: 10.1128/jvi.35.2.519-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T., Vapnek D. Electron microscopic analysis of bacteriophages P1, P1Cm, and P7. Determination of genome sizes, sequence homology, and location of antibiotic-resistance determinants. Virology. 1977 Mar;77(1):376–385. doi: 10.1016/0042-6822(77)90434-2. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]