Abstract
Expression of the multiple interferon-τ (IFN-τ) genes is restricted to embryonic trophectoderm of ruminant ungulate species for a few days in early pregnancy. The promoter regions of these genes are highly conserved. A proximal (bp −91 to −69) sequence has been implicated in controlling trophoblast-specific expression. Here it was used as a target for yeast one-hybrid screening of a day 13 conceptus cDNA library. Two transcription factors of the Ets family, Ets-2 and GABPα, were identified, consistent with the observation that active ovine IFN-τ genes contain a single 10-bp Ets motif (core: GGAA) in the proximal segment, whereas three known inactive ovine genes contain a mutated core motif (TGAA). Cotransfection of a promoter- (−126 to +50) luciferase reporter construct from an active gene (bovineIFN-τ1; boIFNT1) and an Ets-2 expression plasmid in human JAr cells provided up to a 30-fold increase in reporter expression, whereas promoters from inactive genes were not transactivated. GABPα alone was ineffective and had only a ≈2-fold positive effect when coexpressed with its partner GABPβ. Other Ets-related transcription factors, which were not detected in the genetic screen, also provided a range of lesser transactivation effects. Coexpression of Ets-2 and activated Ras failed to transactivate the IFNT promoter greater than Ets-2 alone in JAr cells. The presence of Ets-2 in nuclei of embryonic trophectoderm was confirmed immunocytochemically. Together, these data suggest that Ets-2 plays a role in the transient expression of the nonvirally inducible IFNT genes.
The interferon-τ (IFN-τ) are type I IFN, structurally related to the IFN-β, IFN-α and IFN-ω (with ≈25%, 50%, and 75% primary sequence identities, respectively) (1). Members of this multigene family are expressed within the first epithelium (trophectoderm) of the conceptuses of ruminant ungulate species (cattle, sheep, and their relatives) from the time that of blastocyst hatching to when the trophoblast attaches to the uterine wall. Although exhibiting typical properties of other type I IFN, including an ability to induce an antiviral state in target cells expressing the type I IFN receptor, the function of the IFN-τ is that of preventing the regression of the corpus luteum, an event that normally occurs at the end of the estrous cycle when the animal is not pregnant (1, 2). As a consequence, progesterone production is maintained, and the pregnancy can proceed. This ability of IFN-τ to extend corpus luteum lifespan is achieved indirectly by reducing the pulsatile output of the luteolytic factor, prostaglandin F2α, from the uterine endometrium (1, 2).
The expression of the IFN-τ genes (IFNT) is unusual in at least three respects. First, they are not inducible by virus (3, 4). Second, these genes are transcribed only within the embryonic trophectoderm (5, 6). Third, expression of the IFNT is sustained over several days (1, 5–7) whereas expression of most other type I IFN genes usually occurs in response to virus or other pathogens and is short-lived (8).
The 5′-untranslated regions of the IFNT gene have a high degree of sequence conservation extending up to 400 bp past their transcription start sites (9), but electrophoretic mobility-shift analysis (EMSA) with crude nuclear extract from day 13 ovine (ov) conceptuses has revealed that only two short sequences of DNA beyond the TATA box form stable complexes with putative transcription factors (4). One sequence is proximal (bp −91 to −69 from the transcription start point) and the other more distal (bp −358 to −322). Extracts from more advanced conceptuses, in which IFN-τ expression is absent, provide much more complex and less easily interpretable mobility shift patterns. Transfection experiments have had only limited value because of the lack of appropriate ov or bo cell lines of trophoblast origin that could be transfected. However, transfection studies performed on JAr cells, a human choriocarcinoma cell line, confirmed the importance of the proximal region in control of IFN-τ expression (3, 4), even though such cells, being of human origin, lack genes for the IFN-τ (9, 10).
MATERIALS AND METHODS
Yeast One-Hybrid Screening.
The yeast one-hybrid analysis (11) used the Matchmaker System from CLONTECH. A cDNA library was constructed from day 13 ov conceptus mRNA by using both oligo(dT) and random primers and ligated downstream of the GAL4 activation domain of the pGAD10 yeast expression vector. As the bait, four tandem copies of the promoter sequence (bp −91 to −69, Fig. 1A) from bovine (bo) IFNT1 (12) were inserted into pHISi and pLacZi vectors, which were integrated into Saccharomyces cerevisiae strain, YM4271. Yeast cells transformed with the cDNA library were selected on medium lacking Leu and His and containing 30 mM 3-amino-1,2,4-triazole. Positive colonies generated a blue color on filter papers impregnated with the β-galactosidase (β-gal) substrate (CLONTECH protocol PT1081–1).
Figure 1.
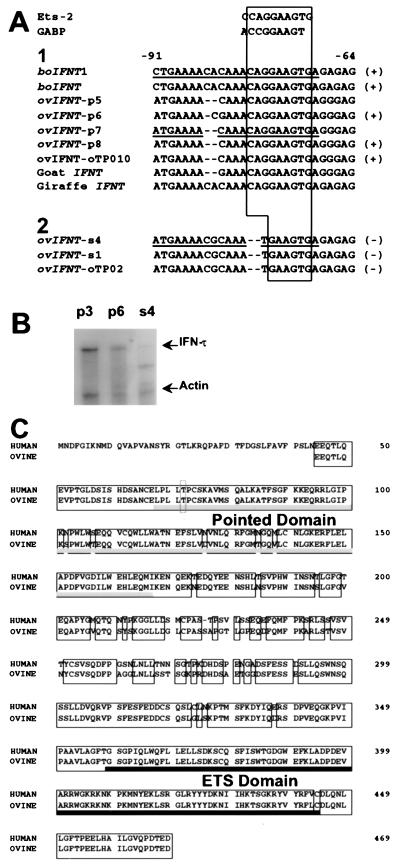
(A) Sequence alignment of the proximal promoter regions of IFNT genes from various species and consensus motifs of Ets-2 (27) and GABP (16). Category 1 sequences represent those genes that contained a conserved Ets motif with an intact GGAA core, whereas category 2 sequences represent genes lacking this conserved motif (core = TGAA). Conserved Ets motif regions are boxed. Genes also identified previously by cDNA library screening (putative transcriptionally active genes) are indicated by (+), and those genes only cloned from genomic DNA (putative transcriptionally inactive genes) are indicated by (−) (14, 29). Underlined sequences represent DNA fragments used for completing EMSA. The references for the sequences are boIFNT1 (12); boIFNT (47); ovIFNT-p7, s4, and s1; and goat IFNT (9), ovIFNT-oTP010 and oTP02 (29); and giraffe IFNT (28). The ovIFNT-p5-, p6-, and p8-promoter sequences have not been reported previously. (B) Relative abundance of specific ov IFN-τ mRNAs in day 12–13 ov conceptuses. RNase protection assays were completed on total cellular RNA (1 μg) from day 12–13 ov conceptuses by using 32P-labeled riboprobes for p3, p6, and s4 ovIFN-τs and for ov actin (14). Full-length protected fragments for all ovIFN-τs (469 bp) and for ov actin (264 bp) were indicated by arrows. Controls, including the unprotected probe in presence and absence of yeast RNA, which showed complete digestion, are not shown. (C) Amino acid sequence comparison of human Ets-2 with Ets-2 identified in the ov cDNA library screen. Conserved sequences are boxed. The Pointed domain (crosshatched bar) and the ETS domain (solid bar) are underlined. The Thr72 residue, which is the site of phosphorylation by Ras-MAP kinase-signaling pathways (20, 36), is indicated by the dashed box. Residues were aligned by computerized alignment software (geneworks; IntelliGenetics, Mountain View, CA).
IFNT-Promoter Sequencing.
The promoter and coding regions of IFNT were isolated from ov liver by PCR (30 cycles; 94°C/50°C/72°C, 1 min each). The primers corresponded to bp −428 to −408 (5′-AATACAAACATCAATATGGCC-3′) from boIFNT1 and bp +677 to +659 (5′-GCGAATTCGCATCTTAGTCGGCAAGAG-3′) from the ovIFNT-p7 gene (13). The 3′-primer sequence is conserved in all identified ovIFNT genes. PCR products (20 total) were subcloned into the pGEM-T easy vector (Promega) and sequenced. The ORF sequences were used to identify the ovIFNT gene variants (4, 13), thereby allowing promoter sequences from all identified clones to be compared.
RNase Protection Assays.
RNase protection assays (14) used 32P-labeled riboprobes antisense to transcripts for ov actin (specific activity = 4.2 × 107 dpm/μg; 264 base-protected fragment) and p3, p6, and s4 ovIFN-τ (average specific activity = 3.6 × 108 dpm/μg; 469 base-protected fragment).
Glutathione S-Transferase (GST) Fusion Proteins.
A 2.4-kb EcoRI fragment of the ov Ets-2 cDNA (encoding amino acids 45–469) was cloned into the EcoRI site of pGEX1 (Amrad, Melbourne, Australia) and used to transform DH5α Escherichia coli. Bacterial growth, IPTG treatment, and preparation of extracts were completed as described (15). The expression of recombinant fusion protein GST–Ets-2 and GST was verified by SDS/PAGE and Western blot analysis by using affinity-purified anti-Ets-2 antibody (Santa Cruz Biotechnology) and alkaline phosphatase conjugated to anti-rabbit IgG (Promega).
EMSA.
DNA-binding reaction mixtures (16) included 4 μg of E. coli protein containing either GST–Ets-2 or GST alone plus 20 fmol of a boIFNT1 (bp −91 to −69) 32P-labeled, double-stranded oligonucleotide probe (≈23,000 cpm). After incubation for 20 min at 25°C, they were analyzed by electrophoresis (16). To verify the specificity of Ets-2 binding, either the indicated competitor DNA (15 pmol) or 0.1 μg of affinity-purified anti-Ets-2 specific antibody, 0.1 μg of anti-mouse IgG or 0.1 μg of normal rabbit serum were added before incubation.
Reporter Gene Constructs and Expression Vectors.
A boIFNT1-promoter fragment (bp −126 to +50) (3) was ligated into the SmaI site of the luciferase-reporter plasmid, pGL2-Basic (Promega), which lacks both eukaryotic promoter and enhancer sequences. The ovIFNT-s4-promoter fragment (bp −126 to +50) (9) was subcloned from pBluescript SK(-) (Stratagene) through KpnI and SacI digestion and inserted into the KpnI/SacI site of pGL2-Basic. Fidelity of constructs was verified by DNA sequencing. The promoter region for mouse urokinase-type plasminogen activator (uPA) also was inserted into the KpnI/HindIII site of pGL2-Basic (17).
The cDNA for human Ets-2 (Ets-2 T72) and Ets-2 with the threonine 72 mutated to alanine (Ets-2 A72) were cloned into the pCGN expression vector (18), which was driven by the cytomegalovirus (CMV) promoter. The activated ras (T24 Ha-ras-1) expression vector pHO6T1 and its parent vector, Homer6, have been described (19). The expression vectors for mouse GABPα and GABPβ1, which were constructed in the CMV promoter-driven expression vector pcDNA1 (Invitrogen), were a gift from M. E. Martin (University of Missouri-Columbia). Additional expression vectors included constructs for chicken Ets-1, mouse Elf-1, human Fli-1, mouse PEA3 (20), and mouse PU.1 (21). The β-gal gene driven by the Rouse sarcoma virus LTR (pRSVLTR-βgal) was a gift from R. V. Guntaka (University of Missouri-Columbia) and was used as an internal control in all transfection experiments.
Cell Culture and Transfection.
JAr cells (3, 4) were plated on 60-mm cell culture dishes (2 × 105 cells/dish) and transfected with 3.2 μg of reporter gene construct DNA and 0.4 μg of expression vector DNA. For transient expression of ras, 1 μg of pHO6T1 was used. For cotransfection with Ets family members, 0.4 μg of expression plasmid was included in CMV-promoter driven Elf-1, Ets-2, and PEA3, whereas 1 μg of expression plasmid was included in Ets-1, PU.1, (SV40 early promoter driven) and Fli-1 (β-actin promoter driven) transfections. All transfections included 50 ng of pRSVLTR-βgal. Total amount of transfected DNA was kept constant in all transfections by including empty vectors. Cells were incubated with DNA/calcium phosphate precipitates in DMEM supplemented with 10% fetal bovine serum for 8 hr. Individual transfections were performed in triplicate and repeated three times or more with at least two different plasmid preparations.
Enzyme Assays for Analysis of Transfection Experiments.
At 36 hr post-transfection, cells were lysed with Galacto-Light Plus lysis solution (Tropix, Bedford, MA). Luciferase activities were determined by injecting luciferase assay reagent (Promega) into cell extracts and recording a 15-s light output from a Turner Designs (Sunnyvale, CA) model 20e luminometer. β-Gal activities were measured by chemiluminesce (15-s light output) after addition of Galacto-Light Plus substrate (Tropix) to cell extracts and after heating to 48°C for 50 min to inactivate endogenous eukaryotic β-gal. The luciferase light units were normalized to β-gal light units, log-transformed to limit heterogeneity of variance, and analyzed by squares analysis of variance (pc-sas, version 6.12, Statistical Analysis System Institute, Cary, NC). Pairwise comparisons among treatments were completed by using F test statistics (pc-sas).
Immunohistochemistry.
Tissue sections (5 μm thick) from paraffin-embedded ov conceptuses (22) were incubated with rabbit affinity-purified anti-Ets-2 specific antibody (Santa Cruz Biotechnology, 100 μg/ml) diluted 1:330 in 1.5% normal goat serum/PBS overnight at 4°C. Binding of Ets-2 antibody to conceptus tissue was detected by using an ABC elite kit and SG substrate kit for peroxidase (Vector Laboratories). Specific binding of anti-Ets-2 IgG was verified by addition of a 1,500-fold molar excess of GST–Ets-2 protein to the anti-Ets-2 IgG solution before exposure to the sections.
RESULTS
Cloning of a Proximal Promoter DNA-Binding Protein.
The day 13 conceptus library was transformed into a yeast strain bearing the tetramerized proximal sequence (bp −91 to −69) (Fig. 1A) of the IFNT gene upstream of both the HIS3- and LacZ-reporter genes. Four colonies from the 2.6 × 106 transformants plated grew on medium lacking Leu and His. All four were capable of transactivating the LacZ-reporter gene. Three of them contained identical sequences, 2,378 bp in length, which appeared to represent the ov equivalent of Ets-2, a transcription factor previously cloned from human and mouse cells (23). The ov sequence lacked the entire 5′-untranslated region as well as the region equivalent to the first 131 nucleotides of the ORF of the human gene (Fig. 1B). Attempts to obtain a full length cDNA from an ov day 13 conceptus cDNA phagemid library (24) also were unsuccessful.
The ov Ets-2 protein was 91% and 87% identical to its human and murine counterparts in the region of overlap. It contained a well conserved pointed domain (Fig. 1C), which had 95% identity with that of human Ets-2. The DNA-binding domain (ETS domain) differed from human Ets-2 at only a single amino acid (a Leu substitution at 443).
The fourth cDNA (1,184 bp) encoded ov GABPα (25), also a member of the Ets family. It, too, was less than full length, encoding a protein 118 residues shorter than mouse GABPα. Sequence identity with human and mouse GABPα, over the overlapping 336 residues was, however, 98% and 95%, respectively, leaving little doubt about its identity.
Proximal Promoter Sequences of IFNTs Relative to Their Expression.
The multimerized proximal sequence used as a bait in the yeast one-hybrid system was from a boIFNT1 (BTP-1 gene) promoter (12) for a gene that is highly expressed in bo embryos (26). It contains a 10-bp sequence (bp −79 to −70) that is identical at nine of ten positions to an Ets-2 consensus sequence (27) (Fig. 1A). There is also a close similarity (identity at 8 of 9 bases) to a sequence known to bind GABPα through its ETS domain (16). In particular, the core motif C/A GGA A/T in the middle of this 10 bp of DNA is characteristic of the majority of sites that interact with Ets family members (27).
The upstream promoters of five ov IFNT and two bo IFNT (including the one described above), whose cDNA have been previously cloned (13, 26) contain the full Ets-2 binding sequence, with an intact GGAA core motif (Fig. 1A). Cloned goat and giraffe genes (9, 28) also contain the same sequence (although these genes have not been proven to be expressed). Importantly, two genes cloned from this laboratory (s4 and s1) (9) and one cloned by Nephew, et al. (29) (oTP-02), whose mRNA transcripts have not been identified in cDNA libraries and may not be highly expressed (14), each have a two base deletion at the 5′ end of the Ets-binding site and lack the core GGAA motif (Fig. 1A). The low expression of s4 is illustrated in Fig. 1B. Whereas strong protected bands are noted for p3 and p6, that for s4 is very faint.
EMSA Analysis of Ets-2 Binding to the IFNT Proximal-Promoter Region.
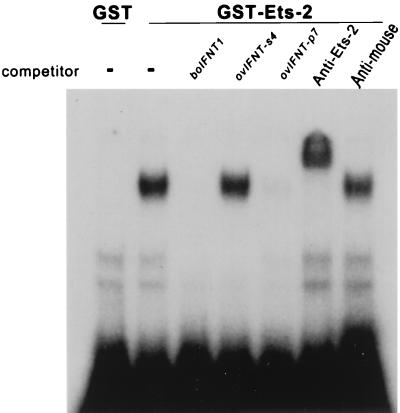
The 32P-labeled proximal-promoter region (bp −91 to −69) of the boIFNT1 gene formed a stable complex with a GST-ov-Ets-2 fusion protein (amino acids 45–469) (Fig. 2). Whereas unlabeled boIFNT1 promoter and the equivalent region of ovIFNT-p7 promoter competed with the radioactive probe for binding to Ets-2, a comparable amount of the equivalent region of ovIFNT-s4 promoter, which lacks the GGAA core sequence (Fig. 1A), did not. A polyclonal antiserum raised against amino acids 450–469 of Ets-2 recognized the putative boIFNT1-Ets-2 complex and gave a supershift. Neither mouse IgG (shown) nor normal rabbit serum (not shown) caused such a supershift.
Figure 2.
DNA binding of ov Ets-2 to the boIFNT proximal-promoter region (bp −91 to −69) and displacement by competitor DNA. E. coli protein containing GST only (negative control) or GST–Ets-2 fusion protein was incubated with 32P-labeled boIFNT1-promoter fragment (Fig. 1A) in presence of either no competitor DNA (−) or 750-fold excess of promoter fragments (boIFNT1, ovIFNT-s4, and p7; Fig. 1A). Additions of anti-Ets-2 antibody (Anti-Ets-2) and anti-mouse IgG (Anti-mouse) to the binding mixture also were tested to verify Ets-2 specificity.
Transactivation of IFNT Promoters by Ets-2 and GABP.
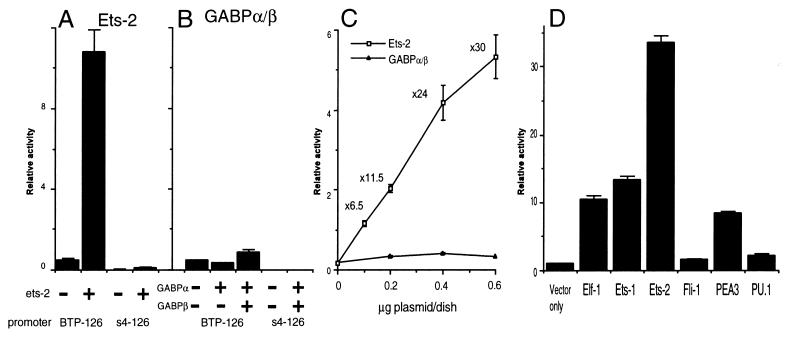
Cotransfection of JAr cells with a luciferase-reporter construct containing the entire 126-bp promoter region of the boIFNT1 gene (BTP-126) and the Ets-2 expression plasmid (pCGN-Ets-2) provided an ≈24-fold transactivation relative to the control. An identical region of the “silent” IFNT gene s4 (bp −126 to +50) (s4–126), placed ahead of the luciferase reporter, was not transactivated by Ets-2.
GABP consists of the Ets-like GABPα subunit, which binds the DNA, and a distinct GABPβ subunit, which provides a transactivation domain to the complex (16, 25). Neither of these subunits alone was able to transactivate the BTP-126- and s4–126-reporter constructs. When both the α and β subunit constructs were transfected together with BTP-126, there was a small induction (≈2-fold) of reporter activity (Fig. 3B). This effect, although slight, was significant (P < 0.05). The s4–126-reporter construct with the mutated Ets response element was not transactivated by the GABPα–GABPβ heterodimer.
Figure 3.
Transactivation of IFNT promoters by human Ets-2, murine GABPα/β and related Ets protens. A boIFNT1 promoter (bp −126 to +50, BTP-126) luciferase reporter and an ovIFNT-s4 promoter (bp −126 to +50, s4–126) luciferase reporter were transfected into human JAr cells in either the absence (−) or the presence (+) of the expression vectors for human Ets-2 (A), murine GABPα, or GABPβ (B). Reporter activity is expressed relative to activity of a cotransfected RSVLTR-βgal plasmid. Results are means (± SE) from three independent experiments. (C) The dose-dependent effects of Ets-2 and GABPα/β transfection on IFNT-promoter activity in human JAr cells. The BTP-126 luciferase construct (3.2 μg) was cotransfected with various amounts (0.1 to 0.6 μg/dish) of human Ets-2 (□) or murine GABPα/β (▴) expression plasmids. After 36 hr, luciferase activity was quantified and normalized for transfection efficiency as measured by β-gal. Results are means (± SE) from three independent experiments. (D) Transactivation of the IFNT promoter by various members of the Ets family proteins. The activity of the BTP-126 luciferase reporter was measured in JAr cells in the presence of Elf-1, Ets-1, Ets-2, Fli-1, PEA3, or PU.1. Results represent means (± SE) from three independent experiments.
The ability of Ets-2 and GABPα/β to transactivate the BTP-126 promoter was examined over a range of transfection concentrations at a fixed concentration of the BTP-126 plasmid (Fig. 3C). Reporter activity was enhanced 6.5-fold by cotransfection with 0.1 μg/60-mm culture dish pCGN-Ets-2 and 30-fold by 0.6 μg/60-mm culture dish. Cotransfection with CMV-GABPα and CMV-GABPβ had only the previously noted minor effect at any of the concentrations tested.
Relative Responsiveness of BTP-126 to Other Ets Family Members.
Several Ets family members, in addition to Ets-2 and GABPα, have been described in mammals, including Ets-1 (23), Elf-1 (30), Fli-1 (31), PEA3 (15). and PU.1 (32). All can bind the core GGAA sequence and appear to regulate a variety of different genes. Their relative ability to transactivate the BTP-126 reporter also varies (Fig. 3D). Whereas Fli-1 and PU.1 were relatively ineffective, the other three (Elf-1, Ets-1, and PEA3) each provided an ≈10-fold increase in reporter activity.
Lack of Ras Respnsiveness of the IFNT (BTP-126) Promoter.
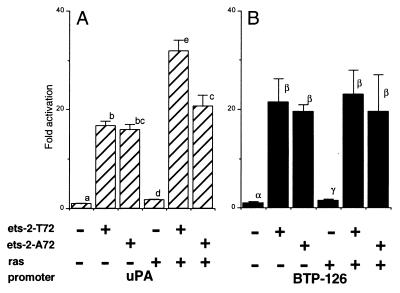
Many cellular genes, including that for uPA (17), contain a Ras-responsive enhancer composed of binding sites for both Ets and AP-1/ATF. In addition, certain Ets family members are direct target for phosphorylation by Ras-mitogen-activated protein (MAP) kinase-signaling pathways (33–35). In the case of Ets-2, the site of phosphorylation and hence of phosphoregulation is Thr72 in the Pointed domain (20, 36). To test whether the IFNT gene promoter is Ras-responsive and whether Ets-2 is a downstream target for the Ras-signaling pathway, wild-type Ets-2 or a form of Ets-2 in which Thr72 had been mutated to Ala were overexpressed in JAr cells in presence of BTP-126 (Fig. 4B). The uPA-reporter gene, which in murine NIH 3T3 cell is activated strongly by the combined expression of Ras and Ets-2 (20), was used as a control. Surprisingly, the uPA promoter was only activated 1.7-fold relative to Ets-2 activation alone when oncogenic Ras and Ets-2 were both overexpressed (Fig. 4A). Nevertheless, this modest effect was statistically significant (P < 0.01) and was not observed with the mutant form Ets-2A72.
Figure 4.
Lack of Ras responsiveness of the IFNT promoter. (A) Activity of the uPA (containing a Ras responsive element, 17)-luciferase reporter, and (B) the IFNT (BTP-126)-luciferase reporter in JAr cells with an expression plasmid for either human Ets-2 (ets-2-T72) or Ets-2 with the threonine 72 mutated to alanine 72 (ets-2-A72). Oncogenic Ras was overexpressed by cotransfectection with an activated Ha-c-Ras expression plasmid. Results are means (± SE) from three independent experiments. Activity is expressed as fold activation to basal. Values marked with different letters within each figure differ significantly (P < 0.01).
Coexpression of Ras and Ets-2 (Ets-2 T72) did not enhance expression from the BTP-126 promoter (Fig. 4B), although there was a tendency for luciferase values to be higher in the Ras/Ets-2 transfected cells. The mutation of Thr72 had no effect.
Localization of Ets-2 in ov Trophoblast.
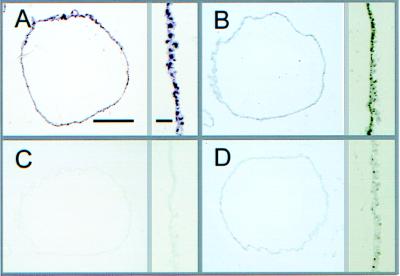
Ets-2 localization was examined by immunocytochemistry in cross-sections of filamentous day 15 ov conceptuses collected when IFN-τ expression is high (5, 22), and the tissue is composed largely of a simple outer epithelial layer of trophectoderm, with a sparse layer of spindle-shaped extraembryonic endoderm loosely attached beneath (Fig. 5A). Immunoperoxidase staining observed after exposing sections to an Ets-2 antiserum gave a strong positive reaction over the nuclei of trophectoderm and a fainter uniform staining over cytoplasmic regions (Fig. 5B). The immunoreactive signal was absent on sections in which the diluted antiserum had first been exposed to GST–Ets-2 fusion protein (Fig. 5C). The specific Ets-2 immunosignal was seen best in Fig. 5D, in which the antiserum had first been adsorbed against GST protein alone.
Figure 5.
Immunolocalization of Ets-2 in cross-sections of filamentous ov conceptuses at day 15 of pregnancy (Bar, 100 μm). A higher magnification view of trophectoderm is shown to the right of each full-frame view (Bar, 10 μm). (A) Hematoxylin/eosin staining. (B) Sections were incubated with the Ets-2 antibody (1:330). (C) The Ets-2 antibody reactivity after adsorption with an excess amount (1,500-fold) of GST–Ets-2 fusion protein. (D) Ets-2 antibody coincubated with an equimolar amount of GST protein.
DISCUSSION
The yeast one-hybrid screening of the day 13 conceptus library provided surprisingly unequivocal results because only two candidate-binding proteins were identified for the proximal promoter element. Both were Ets family members but only one of them, Ets-2, was capable of strong transactivation of the IFN-τ gene promoter in JAr cells. The other, GABPα, was ineffective alone and had only a slight positive effect when coexpressed with its partner, GABPβ. Other Ets-related proteins, which were not detected by the screening procedure, provided a range of transactivation effects.
The IFNT constitute a multigene family, which has been calculated on the basis of nucleotide substitution analyses to have originated by duplication of an IFN-ω gene (IFNW) ≈36 million years ago (10). It has been hypothesized that this event placed the gene into a new genetic context just 5′ the Ets site described here. This change presumably permitted or quickly led to trophoblast expression and directly or indirectly to loss of viral inducibility of the genes. A minimum of four (9) and possibly as many as ten (37) IFNT are present in the genome of sheep and related species. Because these duplications must have occurred relatively recently (10), many of the genes are so similar in sequence that a distinction between individual genes and allelic variants becomes difficult to make. The three “genes” listed in Fig. 1A, which are known to be expressed poorly, if at all, differ sufficiently that they are unlikely to be allelic forms. Each has a deletion and a mutation within the Ets-binding motif that abolished high affinity Ets-2 binding and prevented Ets-2 from transactivating the gene. This mutation seems likely, therefore, to be responsible for the lack of expression of s4 and the related “silent” genes in trophoblast.
Ets family transcription factors regulate a wide variety of genes. However, they have not been previously implicated in controlling type I IFN gene expression. Among the genes known to be activated by Ets-2 are several whose expression is high in epithelial cells, including trophectoderm. They include genes for certain keratins (38), metalloproteinases (39, 40), and the P450 side-chain cleavage enzyme (CYP11A1) (41). In each of these instances, activation by Ets-2 requires a Ras-responsive element accommodating the transcription factor AP1, usually placed close to one (42) or, in some cases, two Ets-binding sites (35, 39). Indeed, most Ets proteins are weak transcriptional activators when acting alone and generally require a coactivator (33). AP1 is a common partner for both Ets-1 and Ets-2 (see refs. 33, 42, and 43 and references therein). In addition to this requirement for a coactivator, Ets-1 and Ets-2 contain a conserved phosphorylatable Thr residue within their Pointed domains, which is a target for Ras acting through the MAP kinase-signaling pathways (20, 36). Presumably only phosphorylated Ets factors interact with AP1 to superactivate the composite enhancer. Although no such involvement of Ras was demonstrated in the activation of the IFNT promoter (BTP-126) in JAr cells (Fig. 4B), Ras, which markedly up-regulates the uPA promoter when coexpressed with Ets-2 in NIH 3T3 cells (20), also had only a modest effect on that promoter in JAr cells. Possibly the Ras-dependent MAP kinase pathway is already operating maximally in these choriocarcinoma cells. Interestingly, the boIFNT1 and other IFNT promoters contain an AP1-like site (TGAGA/GGAG; bp −71 to −64, see Fig. 1A) partially overlapping the Ets site and just proximal to the core GGAA motif. Depending on whether or not these promoters are responsive to Ras activation in embryonic trophectoderm of sheep or cattle, signaling factors that operate through the Ras-MAP kinase pathways might have a role in controlling IFN-τ expression.
Although not ubiquitous, Ets-2 is widely expressed in both adult and embryonic tissues of the mouse (44–46). Therefore, its presence in trophectoderm of sheep (Fig. 5) was not unexpected. Because of its wide tissue distribution, it seems likely that the expression of Ets-2 is not in itself sufficient for IFNT transcription because the IFN-τ are known only to be expressed in trophectoderm (5, 6, 22).
Acknowledgments
We thank Drs. Mark E. Martin and Ramareddy V. Guntaka for expression plasmids and Mr. William A. Ricke for expert assistance with immunohistochemistry.
This work was supported by National Institutes of Health Grant R37HD21896 (to R.M.R.).
ABBREVIATIONS
- IFN-τ
interferon-τ
- IFNT
IFN-τ gene
- bo
bovine
- ov
ovine
- GST
glutathione S-transferase
- MAP
mitogen-activated protein
- EMSA
electrophoretic mobility-shift analysis
- CMV
cytomegalovirus
- β-gal
β-galactosidase
- uPA
urokinase-type plasminogen activator
Footnotes
References
- 1.Roberts R M, Cross J C, Leaman D W. Endocr Rev. 1992;13:432–452. doi: 10.1210/edrv-13-3-432. [DOI] [PubMed] [Google Scholar]
- 2.Bazer F W, Spencer T E, Ott T L. Am J Reprod Immunol. 1997;37:412–420. doi: 10.1111/j.1600-0897.1997.tb00253.x. [DOI] [PubMed] [Google Scholar]
- 3.Cross J C, Roberts R M. Proc Natl Acad Sci USA. 1991;88:3817–3821. doi: 10.1073/pnas.88.9.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leaman D W, Cross J C, Roberts R M. Mol Endocrinol. 1994;8:456–468. doi: 10.1210/mend.8.4.8052267. [DOI] [PubMed] [Google Scholar]
- 5.Farin C E, Imakawa K, Hansen T R, McDonnell J J, Murphy C N, Farin P W, Roberts R M. Biol Reprod. 1990;43:210–218. doi: 10.1095/biolreprod43.2.210. [DOI] [PubMed] [Google Scholar]
- 6.Guillomot M, Michel C, Gaye P, Charlier M, Trojan J, Martal J. Biol Cell. 1990;68:205–211. doi: 10.1016/0248-4900(90)90309-q. [DOI] [PubMed] [Google Scholar]
- 7.Hansen T R, Imakawa K, Polites H G, Marotti K R, Anthony R V, Roberts R M. J Biol Chem. 1988;263:3060–3067. [PubMed] [Google Scholar]
- 8.De Maeyer E M, De Maeyer-Gignard J. Interferons and Other Regulatory Cytokines. New York: Wiley Interscience; 1988. [Google Scholar]
- 9.Leaman D W, Roberts R M. J Interferon Res. 1992;12:1–11. doi: 10.1089/jir.1992.12.1. [DOI] [PubMed] [Google Scholar]
- 10.Roberts R M, Liu L, Alexenko A. Prog Nucleic Acid Res Mol Biol. 1997;56:287–325. doi: 10.1016/s0079-6603(08)61008-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang M M, Reed R R. Nature (London) 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 12.Hansen T R, Leaman D W, Cross J C, Mathaialagan N, Bixby J A, Roberts R M. J Biol Chem. 1991;266:3060–3067. [PubMed] [Google Scholar]
- 13.Klemann S W, Imakawa K, Roberts R M. Nucleic Acids Res. 1990;18:6724. doi: 10.1093/nar/18.22.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ealy A D, Green J A, Alexenko A P, Keisler D H, Roberts R M. Biol Reprod. 1998;58:566–573. doi: 10.1095/biolreprod58.2.566. [DOI] [PubMed] [Google Scholar]
- 15.Xin J-H, Cowie A, Lachance P, Hassell J A. Genes Dev. 1992;6:481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]
- 16.Brown T A, McKnight S L. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 17.Stacey K J, Fowles L F, Colman M S, Ostrowski M C, Hume D A. Mol Cell Biol. 1995;15:3430–3441. doi: 10.1128/mcb.15.6.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka M, Herr W. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 19.Spandidos D A, Wilkie N M. Nature (London) 1984;310:469–475. doi: 10.1038/310469a0. [DOI] [PubMed] [Google Scholar]
- 20.Yang B-S, Hauser C A, Henkel G, Coleman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy M A, Yang B-S, Yue X, Barnett C J, Ross I L, Sweet M J, Hume D A, Ostrowski M C. J Exp Med. 1994;180:2309–2319. doi: 10.1084/jem.180.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farin C E, Imakawa K, Roberts R M. Mol Endocrinol. 1989;3:1099–1107. doi: 10.1210/mend-3-7-1099. [DOI] [PubMed] [Google Scholar]
- 23.Watson D K, McWilliams M J, Lapis P, Lautenberger J A, Schweinfest C W, Papas T S. Proc Natl Acad Sci USA. 1988;85:7862–7866. doi: 10.1073/pnas.85.21.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer K K, Duffy J Y, Klemann S W, Bixby J A, Low B G, Pope W F, Roberts R M. J Biol Chem. 1994;269:7255–7261. [PubMed] [Google Scholar]
- 25.Thompson C C, Brown T A, McKnight S L. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- 26.Imakawa K, Hansen T R, Malathy P-V, Anthony R V, Polites H G, Marotti K R, Roberts R M. Mol Endocrinol. 1989;3:127–139. doi: 10.1210/mend-3-1-127. [DOI] [PubMed] [Google Scholar]
- 27.Wasylyk B, Hahn S L, Giovane A. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Leaman D W, Roberts R M. J Interferon Res. 1996;16:949–951. doi: 10.1089/jir.1996.16.949. [DOI] [PubMed] [Google Scholar]
- 29.Nephew K P, Whaley A E, Christenson R K, Imakawa K. Biol Reprod. 1993;48:768–778. doi: 10.1095/biolreprod48.4.768. [DOI] [PubMed] [Google Scholar]
- 30.Thompson C B, Wang C-Y, Ho I-C, Bohjanen P R, Petryniak B, June C H, Miesfeldt S, Zhang L, Nabel G, Karpinski B, et al. Mol Cell Biol. 1992;12:1043–1053. doi: 10.1128/mcb.12.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-David Y, Giddens E B, Letwin K, Bernstain A. Genes Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 32.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 33.Crepieux P, Coll J, Stehelin D. Crit Rev Oncog. 1994;5:615–638. [PubMed] [Google Scholar]
- 34.de Launoit Y, Baert J-L, Chotteau A, Monte D, Defossez P-A, Coutte L, Pelczar H, Leenders F. Biochem Mol Med. 1997;61:127–135. doi: 10.1006/bmme.1997.2605. [DOI] [PubMed] [Google Scholar]
- 35.Galang C K, Der C J, Hauser C A. Oncogene. 1994;9:2913–2921. [PubMed] [Google Scholar]
- 36.McCarthy S A, Chen D, Yang B-S, Garcia Ramires J J, Cherwinski H, Chen X-R, Klagsbrun M, Hauser C A, Ostrowski M C, McMahon M. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan A M, Womack J E. Anim Genet. 1993;24:9–16. doi: 10.1111/j.1365-2052.1993.tb00912.x. [DOI] [PubMed] [Google Scholar]
- 38.Oshima R G, Baribault H, Caulin C. Cancer Metastasis Rev. 1996;15:445–471. doi: 10.1007/BF00054012. [DOI] [PubMed] [Google Scholar]
- 39.Buttice G, Kurkinen M. J Biol Chem. 1993;268:7196–7204. [PubMed] [Google Scholar]
- 40.Borden P, Heller R A. Crit Rev Eukaryotic Gene Expression. 1997;7:159–178. doi: 10.1615/critreveukargeneexpr.v7.i1-2.90. [DOI] [PubMed] [Google Scholar]
- 41.Pestell R G, Albanese C, Watanabe G, Lee R J, Lastowiecki P, Zon L, Ostrowski M, Jameson J L. Mol Endocrinol. 1996;10:1084–1094. doi: 10.1210/mend.10.9.8885243. [DOI] [PubMed] [Google Scholar]
- 42.Reddy M A, Langer S J, Colman M S, Ostrowski M C. Mol Endocrinol. 1992;6:1051–1060. doi: 10.1210/mend.6.7.1324418. [DOI] [PubMed] [Google Scholar]
- 43.Wasylyk B, Wasylyk C, Flores P, Begue A, Leprince D, Stehelin D. Nature (London) 1990;346:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 44.Bhat N K, Fisher R J, Fujiwara S, Ascione R, Papas T K. Proc Natl Acad Sci USA. 1987;84:3161–3165. doi: 10.1073/pnas.84.10.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kola I, Brookes S, Green A R, Garber R, Tymms M, Papas T S, Seth A. Proc Natl Acad Sci USA. 1993;90:7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maroulakou I G, Papas T S, Green J E. Oncogene. 1994;9:1551–1565. [PubMed] [Google Scholar]
- 47.Stewart H J, McCann S H E, Flint A P F. J Mol Endocrinol. 1990;4:275–282. doi: 10.1677/jme.0.0040275. [DOI] [PubMed] [Google Scholar]