Abstract
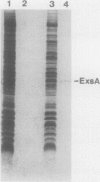
Exoenzyme S is an ADP-ribosyltransferase enzyme distinct from exotoxin A that is synthesized and secreted by Pseudomonas aeruginosa. Yields of exoenzyme S are variable and depend on strain and growth conditions. Since certain medium additives are required for exoenzyme S production, its regulation may be influenced by environmental stimuli. In this study, we have cloned a region that complements the exoenzyme S-deficient phenotype of strain 388 exs1::Tn1, a chromosomal Tn1 insertional mutation. A large clone (28 kb) was shown to restore both synthesis and secretory functions to the mutant strain. Subcloning and Tn501 mutagenesis experiments localized the region required for exoenzyme S synthesis to a 3.2-kb fragment. Nucleotide sequence analysis demonstrated several open reading frames. Comparison of the N-terminal amino acid sequence of purified exoenzyme S with predicted amino acid sequences of all open reading frames indicated that the structural gene was not encoded within the sequenced region. Homology studies suggested that the region encoded three regulatory genes, exsC, exsB, and exsA. ExsA was homologous to the AraC family of transcriptional activator proteins, with extensive homology being found with one member of this family, VirF of Yersinia enterocolitica. VirF and ExsA both contain carboxy-terminal domains with the helix-turn-helix motif of DNA-binding proteins. The ExsA gene product appeared to be required for induction of exoenzyme S synthesis above a low basal level. Expression of ExsA was demonstrated by cloning the region under the control of the T7 promoter. Gene replacement experiments suggested that the expression of ExsC affects the final yield of exoenzyme S.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjorn M. J., Pavlovskis O. R., Thompson M. R., Iglewski B. H. Production of exoenzyme S during Pseudomonas aeruginosa infections of burned mice. Infect Immun. 1979 Jun;24(3):837–842. doi: 10.1128/iai.24.3.837-842.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Coburn J., Dillon S. T., Iglewski B. H., Gill D. M. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989 Mar;57(3):996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn J., Wyatt R. T., Iglewski B. H., Gill D. M. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989 May 25;264(15):9004–9008. [PubMed] [Google Scholar]
- Cornelis G. R., Biot T., Lambert de Rouvroit C., Michiels T., Mulder B., Sluiters C., Sory M. P., Van Bouchaute M., Vanooteghem J. C. The Yersinia yop regulon. Mol Microbiol. 1989 Oct;3(10):1455–1459. doi: 10.1111/j.1365-2958.1989.tb00129.x. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Sluiters C., de Rouvroit C. L., Michiels T. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989 Jan;171(1):254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Grimwood K., To M., Rabin H. R., Woods D. E. Inhibition of Pseudomonas aeruginosa exoenzyme expression by subinhibitory antibiotic concentrations. Antimicrob Agents Chemother. 1989 Jan;33(1):41–47. doi: 10.1128/aac.33.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Feramisco J. R., Fiddes J. C., Thomas G. P., Hughes S. H. Identification of clones that encode chicken tropomyosin by direct immunological screening of a cDNA expression library. Proc Natl Acad Sci U S A. 1983 Jan;80(1):31–35. doi: 10.1073/pnas.80.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Morgan A. F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979 Mar;43(1):73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglewski B. H., Sadoff J., Bjorn M. J., Maxwell E. S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Bradley J., Lochner J. E., Iglewski B. H. The role of exoenzyme S in infections with Pseudomonas aeruginosa. J Infect Dis. 1985 Oct;152(4):716–721. doi: 10.1093/infdis/152.4.716. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Iglewski B. H. Contribution of exoenzyme S to the virulence of Pseudomonas aeruginosa. Antibiot Chemother (1971) 1985;36:40–48. doi: 10.1159/000410470. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Iglewski B. H. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984 Aug;45(2):470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicas T. I., Iglewski B. H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985 Apr;31(4):387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- Ohman D. E., West M. A., Flynn J. L., Goldberg J. B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol P. A., Dennis J. J., MacDougall P. C., Sexton M., Woods D. E. Cloning and expression of the Pseudomonas aeruginosa exoenzyme S toxin gene. Microb Pathog. 1990 Apr;8(4):243–257. doi: 10.1016/0882-4010(90)90051-q. [DOI] [PubMed] [Google Scholar]
- Staudenmaier H., Van Hove B., Yaraghi Z., Braun V. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J Bacteriol. 1989 May;171(5):2626–2633. doi: 10.1128/jb.171.5.2626-2633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F. F., Klee H. J., Nester E. W. In vivo packaging of cosmids in transposon-mediated mutagenesis. J Bacteriol. 1983 Feb;153(2):1075–1078. doi: 10.1128/jb.153.2.1075-1078.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. E., Iglewski B. H. Toxins of Pseudomonas aeruginosa: new perspectives. Rev Infect Dis. 1983 Sep-Oct;5 (Suppl 4):S715–S722. doi: 10.1093/clinids/5.supplement_4.s715. [DOI] [PubMed] [Google Scholar]
- Woods D. E., Sokol P. A. Use of transposon mutants to assess the role of exoenzyme S in chronic pulmonary disease due to Pseudomonas aeruginosa. Eur J Clin Microbiol. 1985 Apr;4(2):163–169. doi: 10.1007/BF02013591. [DOI] [PubMed] [Google Scholar]
- Woods D. E., To M., Sokol P. A. Pseudomonas aeruginosa exoenzyme S as a pathogenic determinant in respiratory infections. Antibiot Chemother (1971) 1989;42:27–35. doi: 10.1159/000417600. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]