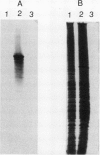
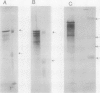
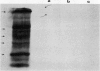
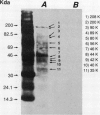
Abstract
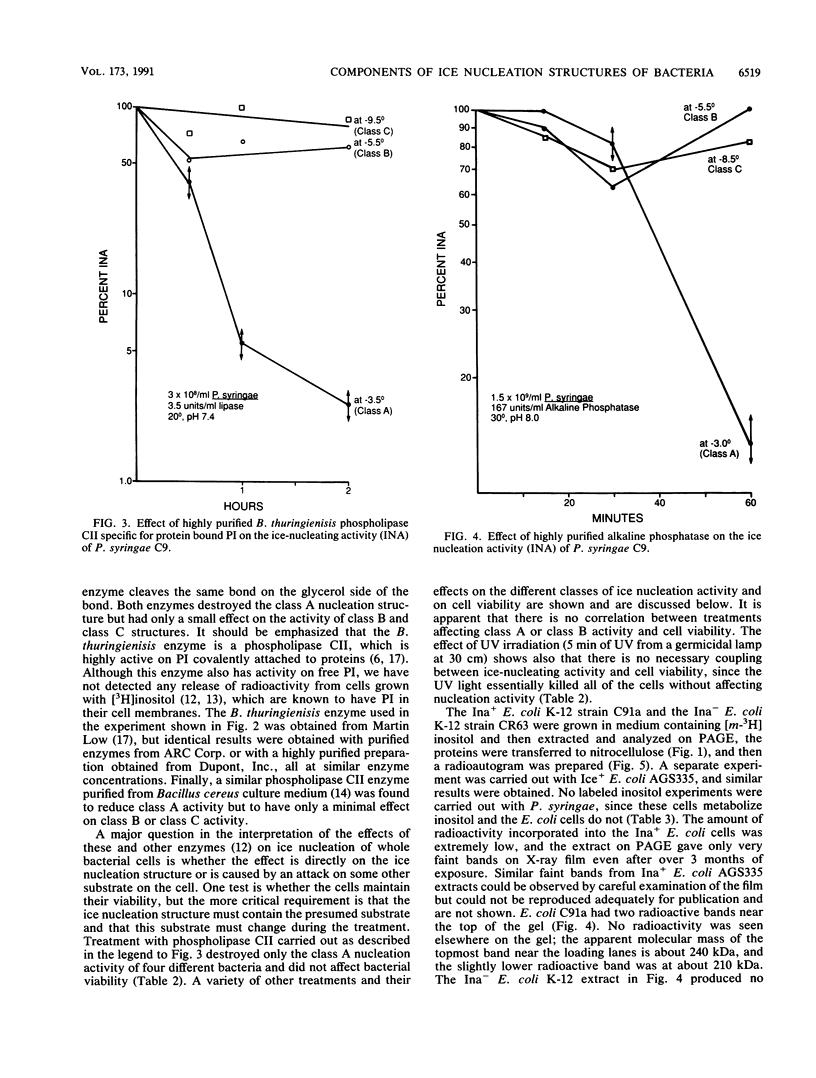
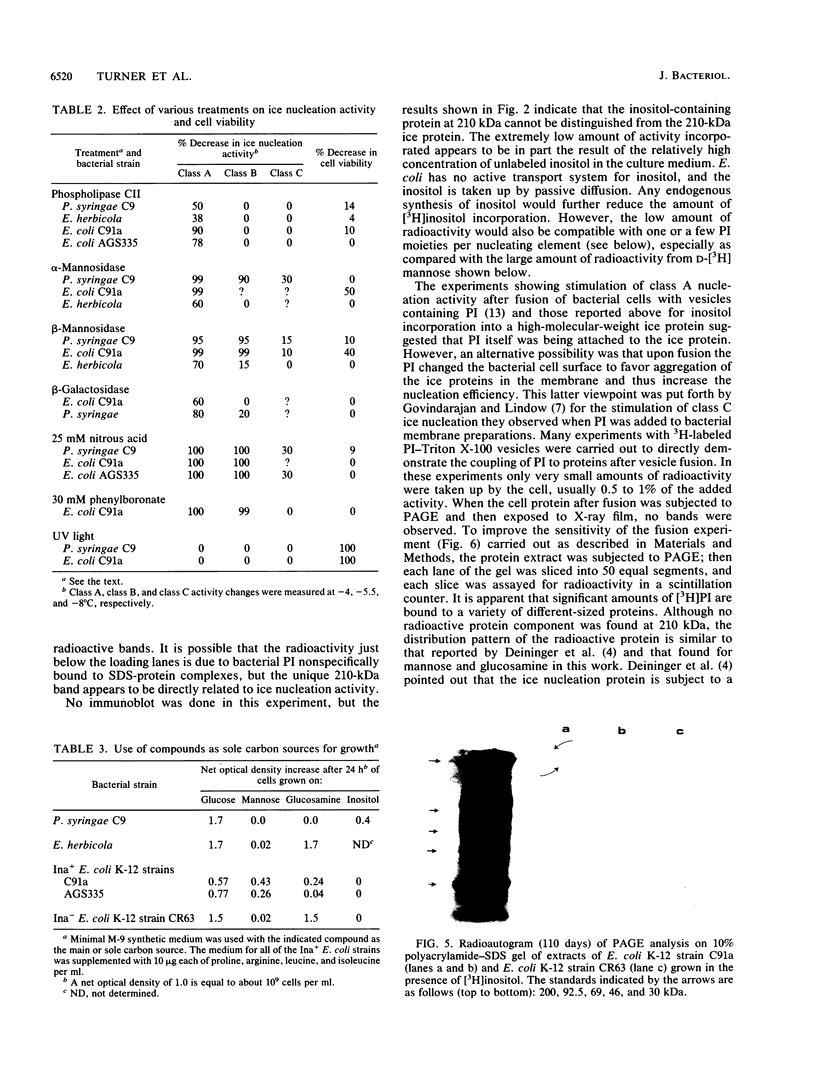
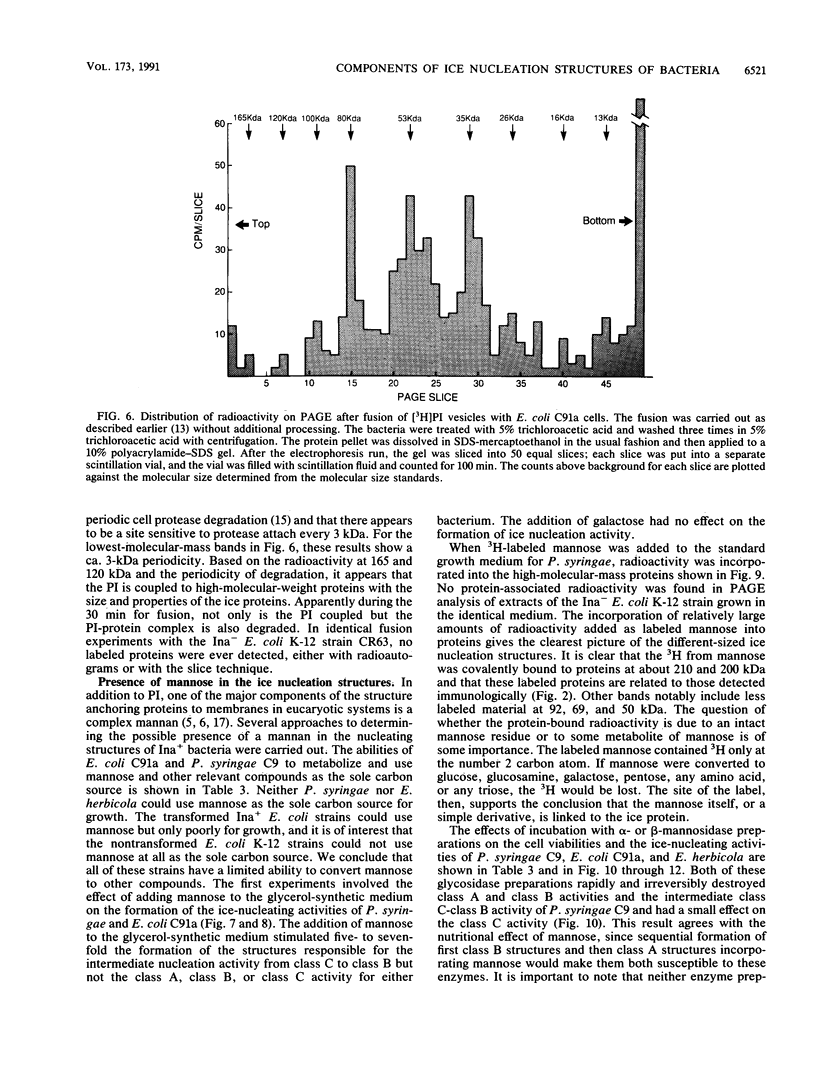
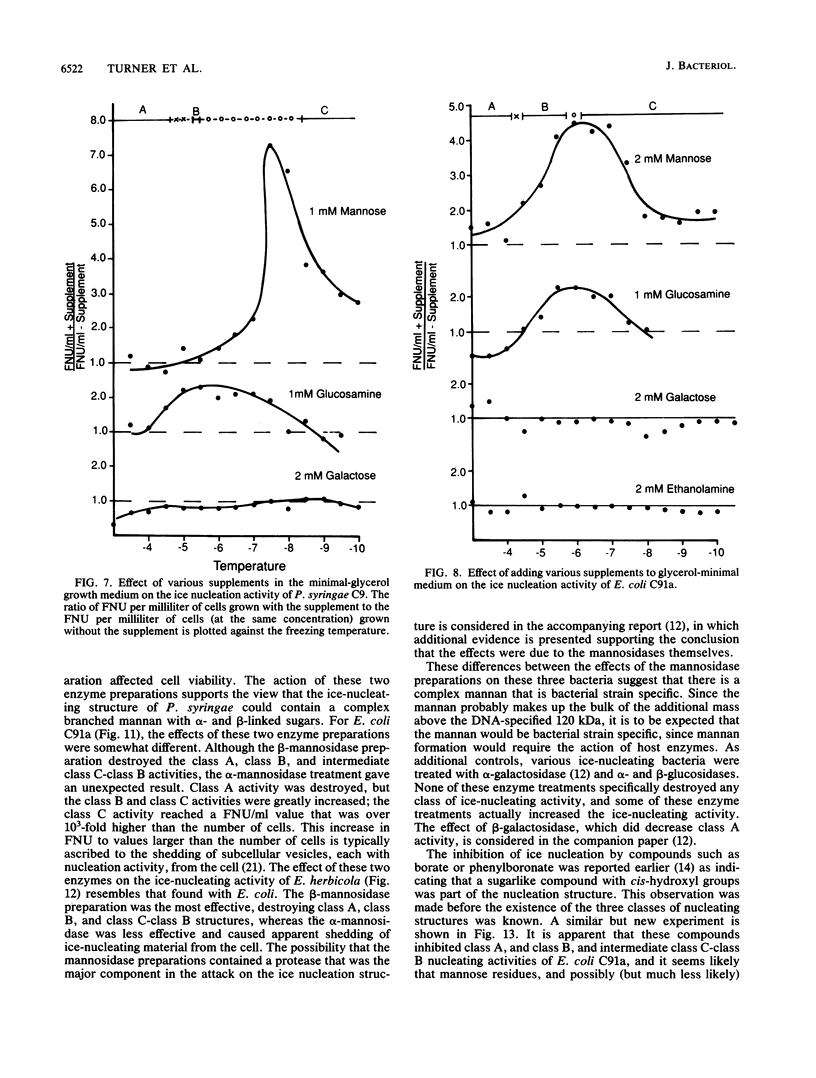
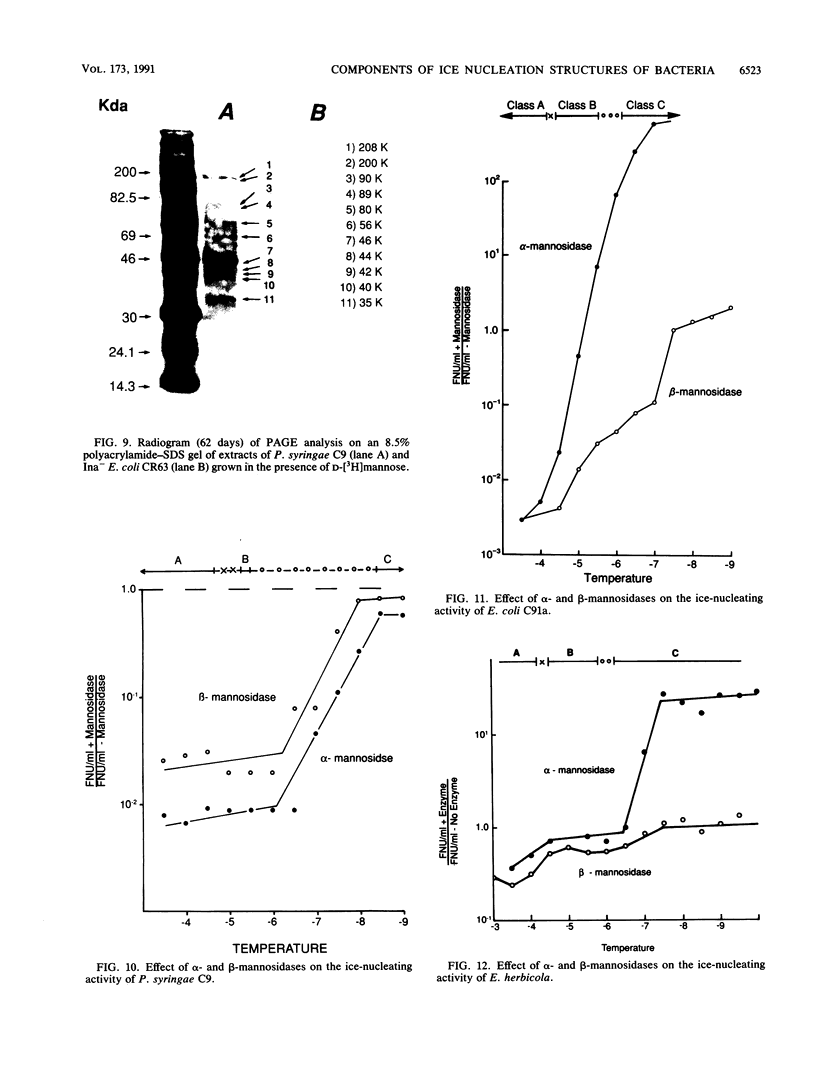
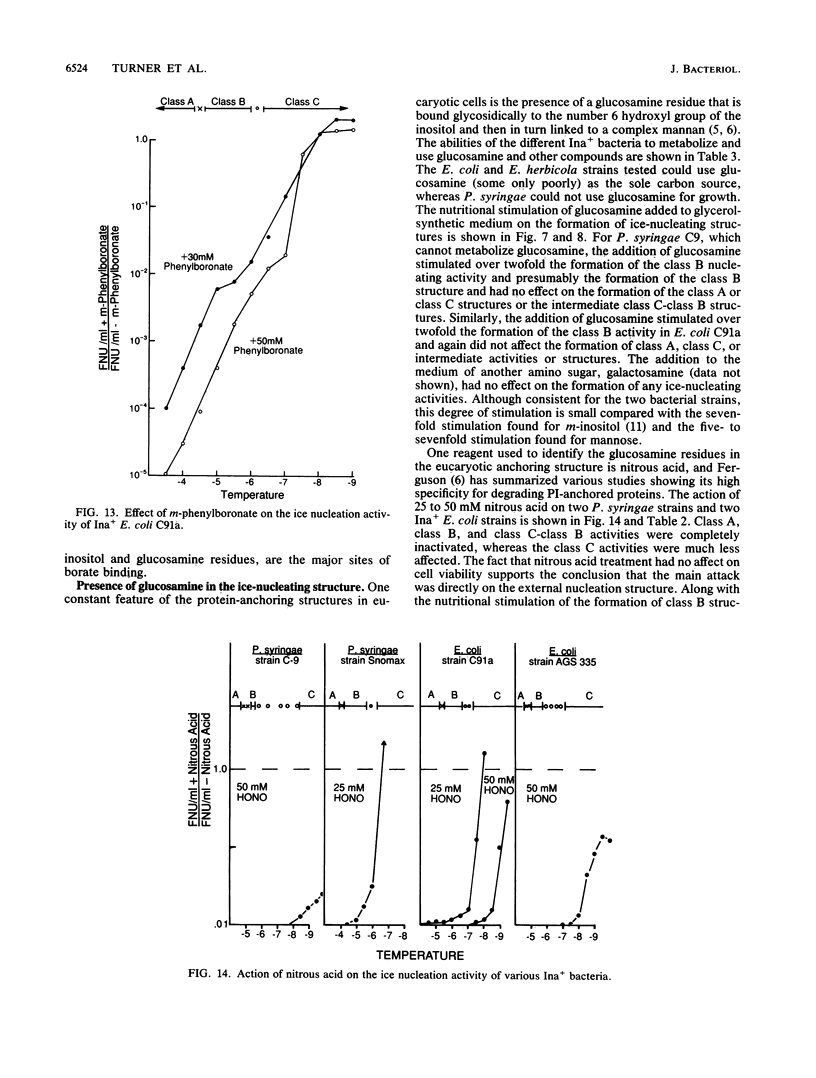
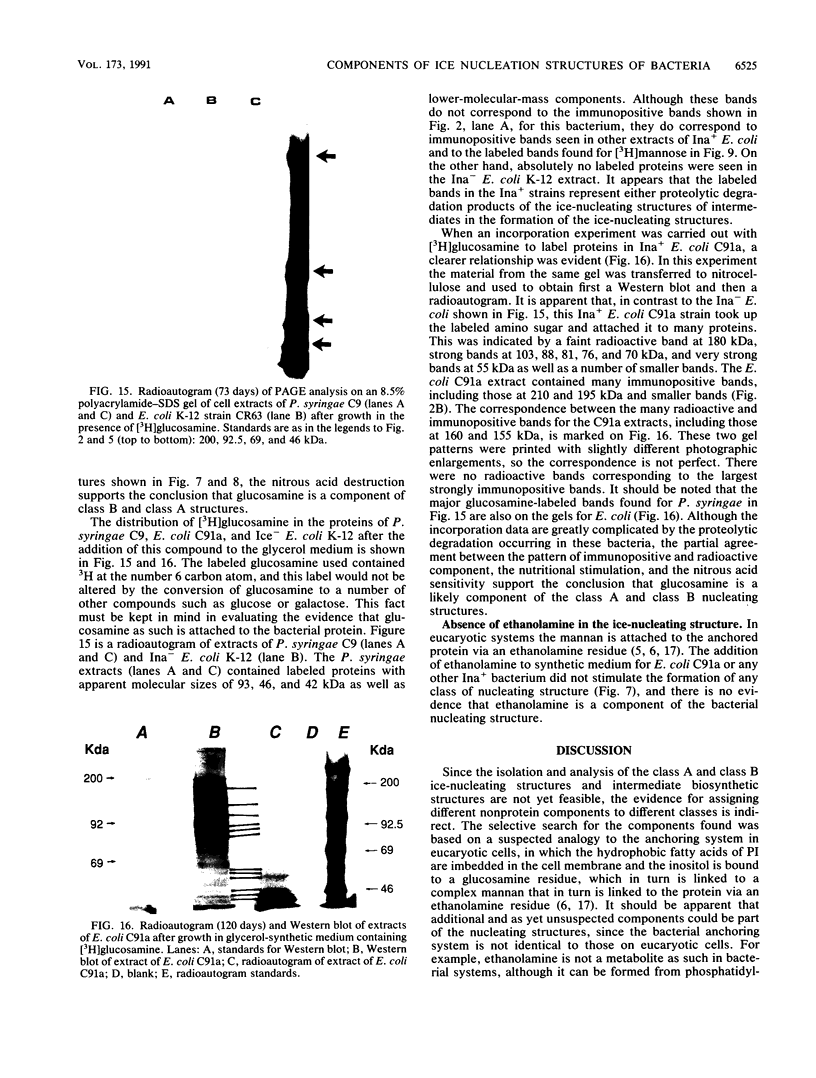
Nonprotein components attached to the known protein product of the inaZ gene of Pseudomonas syringae have been identified and shown to be necessary for the most efficient ice nucleation of supercooled H2O. Previous studies have shown that cultures of Ina+ bacteria have cells with three major classes of ice-nucleating structures with readily differentiated activities. Further, some cells in the culture have nucleating activities intermediate between those of the different classes and presumably have structures that are biosynthetic intermediates between those of the different classes. Since these structures cannot be readily isolated and analyzed, their components have been identified by the use of specific enzymes or chemical probes, by direct incorporation of labeled precursors, and by stimulation of the formation of specific classes of freezing structures by selective additions to the growth medium. From these preliminary studies it appears that the most active ice nucleation structure (class A) contains the ice nucleation protein linked to phosphatidylinositol and mannose, probably as a complex mannan, and possibly glucosamine. These nonprotein components are characteristic of those used to anchor external proteins to cell membranes of eucaryotic cells and suggest that a similar but not identical anchoring mechanism is required for efficient ice nucleation structure. The class B structure has been found to contain protein presumably linked to the mannan and glucosamine moieties but definitely not to the phosphatidylinositol. The class C structure, which has the poorest ice nucleation activity, appears to be the ice nucleation protein linked to a few mannose residues and to be partially imbedded in the outer cell membrane.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Corotto L. V., Wolber P. K., Warren G. J. Ice nucleation activity of Pseudomonas fluorescens: mutagenesis, complementation analysis and identification of a gene product. EMBO J. 1986 Feb;5(2):231–236. doi: 10.1002/j.1460-2075.1986.tb04203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
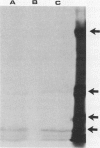
- Deininger C. A., Mueller G. M., Wolber P. K. Immunological characterization of ice nucleation proteins from Pseudomonas syringae, Pseudomonas fluorescens, and Erwinia herbicola. J Bacteriol. 1988 Feb;170(2):669–675. doi: 10.1128/jb.170.2.669-675.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering T. L., Masterson W. J., Hart G. W., Englund P. T. Biosynthesis of glycosyl phosphatidylinositol membrane anchors. J Biol Chem. 1990 Jan 15;265(2):611–614. [PubMed] [Google Scholar]
- Ferguson M. A., Williams A. F. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu Rev Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- Govindarajan A. G., Lindow S. E. Size of bacterial ice-nucleation sites measured in situ by radiation inactivation analysis. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1334–1338. doi: 10.1073/pnas.85.5.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Yokoyama S., Calhoun D. H. Role of cryptic genes in microbial evolution. Mol Biol Evol. 1983 Dec;1(1):109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M., Westaway D. Phosphatidylinositol as a Component of the Ice Nucleating Site of Pseudomonas syringae and Erwinia herbiola. Science. 1984 Nov 16;226(4676):845–846. doi: 10.1126/science.226.4676.845. [DOI] [PubMed] [Google Scholar]
- Kozloff L. M., Schofield M. A., Lute M. Ice nucleating activity of Pseudomonas syringae and Erwinia herbicola. J Bacteriol. 1983 Jan;153(1):222–231. doi: 10.1128/jb.153.1.222-231.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Turner M. A., Arellano F. Formation of bacterial membrane ice-nucleating lipoglycoprotein complexes. J Bacteriol. 1991 Oct;173(20):6528–6536. doi: 10.1128/jb.173.20.6528-6536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Turner M. A., Arellano F., Lute M. Phosphatidylinositol, a phospholipid of ice-nucleating bacteria. J Bacteriol. 1991 Mar;173(6):2053–2060. doi: 10.1128/jb.173.6.2053-2060.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppe A., Evans L. M., McMillen D. A., Griffith O. H. Phosphatidylinositol-specific phospholipase C of Bacillus cereus: cloning, sequencing, and relationship to other phospholipases. J Bacteriol. 1989 Nov;171(11):6077–6083. doi: 10.1128/jb.171.11.6077-6083.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore F. S., Waxman L., Goldberg A. L. Studies of the ATP-dependent proteolytic enzyme, protease La, from Escherichia coli. J Biol Chem. 1982 Apr 25;257(8):4187–4195. [PubMed] [Google Scholar]
- Low M. G. Glycosyl-phosphatidylinositol: a versatile anchor for cell surface proteins. FASEB J. 1989 Mar;3(5):1600–1608. doi: 10.1096/fasebj.3.5.2522071. [DOI] [PubMed] [Google Scholar]
- Mueller G. M., Wolber P. K., Warren G. J. Clustering of ice nucleation protein correlates with ice nucleation activity. Cryobiology. 1990 Aug;27(4):416–422. doi: 10.1016/0011-2240(90)90018-y. [DOI] [PubMed] [Google Scholar]
- Phelps P., Giddings T. H., Prochoda M., Fall R. Release of cell-free ice nuclei by Erwinia herbicola. J Bacteriol. 1986 Aug;167(2):496–502. doi: 10.1128/jb.167.2.496-502.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth M. W., Wolber P. K., Warren G. J. Nonlinear relationship between concentration and activity of a bacterial ice nucleation protein. J Biol Chem. 1988 Oct 15;263(29):15211–15216. [PubMed] [Google Scholar]
- Tinker D., Rucker R. B. Role of selected nutrients in synthesis, accumulation, and chemical modification of connective tissue proteins. Physiol Rev. 1985 Jul;65(3):607–657. doi: 10.1152/physrev.1985.65.3.607. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. A., Arellano F., Kozloff L. M. Three separate classes of bacterial ice nucleation structures. J Bacteriol. 1990 May;172(5):2521–2526. doi: 10.1128/jb.172.5.2521-2526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G., Corotto L. The consensus sequence of ice nucleation proteins from Erwinia herbicola, Pseudomonas fluorescens and Pseudomonas syringae. Gene. 1989 Dec 21;85(1):239–242. doi: 10.1016/0378-1119(89)90488-5. [DOI] [PubMed] [Google Scholar]
- Wolber P. K., Deininger C. A., Southworth M. W., Vandekerckhove J., van Montagu M., Warren G. J. Identification and purification of a bacterial ice-nucleation protein. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7256–7260. doi: 10.1073/pnas.83.19.7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber P., Warren G. Bacterial ice-nucleation proteins. Trends Biochem Sci. 1989 May;14(5):179–182. doi: 10.1016/0968-0004(89)90270-3. [DOI] [PubMed] [Google Scholar]