Abstract
Transporters in the Golgi apparatus membrane translocate nucleotide sugars from the cytosol into the Golgi lumen before these can be substrates for the glycosylation of proteins, lipids, and proteoglycans. We have cloned the mammalian Golgi membrane transporter for uridine diphosphate-N-acetylglucosamine by phenotypic correction with cDNA from MDCK cells of a recently characterized Kluyveromyces lactis mutant deficient in Golgi transport of the above nucleotide sugar. Phenotypically corrected transformants were separated from mutants in a fluorescent-activated cell sorter after labeling of K. lactis cells with fluorescein isothiocyanate (FITC) conjugated to Griffonia simplicifolia II lectin, which binds terminal N-acetylglucosamine. A 2-kb DNA fragment was found to restore the wild-type cell lectin binding phenotype, which reverted to the mutant one upon loss of the plasmid. The DNA fragment contained an ORF encoding a hydrophobic, multitransmembrane spanning protein of 326 aa that had only 22% amino acid sequence identity with the corresponding transporter from K. lactis but showed 53% amino acid sequence identity to the mammalian UDP-galactose transporters and 40% to the CMP-sialic acid transporter. Golgi vesicles from the transformant regained their ability to transport UDP-GlcNAc in an assay in vitro. The above results demonstrate that the mammalian Golgi UDP-GlcNAc transporter gene has all of the necessary information for the protein to be expressed and targeted functionally to the Golgi apparatus of yeast and that two proteins with very different amino acid sequences may transport the same solute within the same Golgi membrane.
Keywords: membranes/glycoproteins/proteoglycans/glycosylation
Transport of UDP-N-acetylglucosamine across the Golgi membrane is required for this nucleotide sugar to enter the lumen of the Golgi apparatus from its site of synthesis in the cytosol; in the lumen, UDP-GlcNAc is a substrate in the biosynthesis of glycoproteins and proteoglycans such as heparin and heparan sulfate (1). Transporters of different nucleotide sugars in the Golgi membrane of mammalian cells, yeast, and protozoa recently have been cloned by phenotypic correction of mutants in these activities (1). The transporters that are antiporters are hydrophobic, multitransmembrane spanning proteins (1). Studies in yeast and mammalian cells suggest that these proteins regulate posttranslational modifications in the Golgi lumen through the availability of nucleotide sugars in this compartment (1–3).
We recently described a mutant of Kluyveromyces lactis, which is specifically deficient in transport of uridine diphosphate N-acetylglucosamine into its Golgi lumen (4). The mutant lacks N-acetylglucosamine in its outer mannan chains (5). By phenotypic complementation of this mutant with K. lactis genomic DNA, a hydrophobic, multitransmembrane spanning protein of 328 aa was identified; upon transformation, Golgi vesicles from the mutant regained their ability to transport UDP-N-acetylglucosamine (6).
Comparison of amino acid sequences between the transporters of UDP-N-acetylglucosamine from K. lactis and UDP-galactose from humans (7) showed only 22% of identity. This result was surprising, particularly in view of the fact that both uridine nucleotide sugar transporters use UMP as antiporters and transport of both nucleotide sugars is inhibited competitively by uridine mono- and diphosphates (8). This result and our inability to observe even at low stringency a homologous RNA in mammalian cells, using the entire K. lactis UDP-GlcNAc transporter gene DNA as a probe, led us to attempt to clone the mammalian Golgi UDP-N-acetylglucosamine transporter by phenotypic complementation of the K. lactis UDP-GlcNAc transporter mutant with the cDNA of MDCK cells.
We recently showed that MDCK cells synthesize, in addition to glycoproteins and glycosphingolipids, keratan sulfate, chondroitin sulfate, and heparan sulfate (3). A ricin-resistant, mutant MDCK cell line, 95% defective in transport of UDP-galactose into the Golgi lumen, was deficient in the biosynthesis of galactose containing glycoproteins, glycosphingolipids, and keratan sulfate (which contains galactose in its glycosaminoglycan polymer) but surprisingly not in chondroitin sulfate and heparan sulfate, both of which contain galactose solely in their linkage region (3, 9). This is the first evidence that limited supply of nucleotide sugars selectively affects the biosynthesis of macromolecules in the Golgi lumen of mammalian cells and extends our previous observations in Saccharomyces cerevisiae, in which we showed that, upon limited Golgi luminal supply of GDP-mannose, differential mannosylation of proteins occurred (2).
K. lactis cells deficient in transport of UDP-GlcNAc were transformed with a cDNA library from MDCK cells under control of the yeast constitutive phosphoglycero kinase (PGK) promoter in a pKD1-derived plasmid. Cells were then labeled with fluorescein isothiocyanate conjugated to Griffonia simplicifolia II (GSII) lectin, which recognizes terminal α- or β-linked N-acetylglucosamine. We previously showed that mutant and wild-type cells can be separated in a fluorescent-activated cell sorter (FACS) after labeling of cells with this lectin (6). After several rounds of cell sorting, a 2-kb DNA fragment was isolated, sequenced, and found to encode an ORF for a multitransmembrane-spanning, 326-aa protein. Upon loss of the plasmid containing this fragment from the transformant, reversion to the mutant phenotype occurred. In an assay in vitro, Golgi vesicles from the transformant had regained their ability to transport UDP-GlcNAc into their lumen. Together, the above results demonstrate that (i) the Golgi UDP-GlcNAc transporter protein of the MDCK cells has the same function as the corresponding one from K. lactis even though they share very little amino acid sequence identity and (ii) a mammalian Golgi transporter protein has the necessary Golgi targeting information to allow it to be expressed in a functional manner in a heterologous system such as yeast and thus result in phenotypic correction of a cell surface defect.
MATERIALS AND METHODS
[α-32P]dCTP (3,000 Ci/mmol) and UDP-[3H]GlcNAc (29 Ci/mmol) were purchased from DuPont/NEN. GDP[3H]Man (15 Ci/mmol) was purchased from American Radiochemicals. GSII lectin conjugated to FITC (GSII-FITC) was purchased from EY Laboratories. Restriction enzymes were purchased from New England Biolabs.
Strains, Media, and Growth Conditions.
The following K. lactis strains were used in this study: KL3 (Mat a, uraA, mnn2–2, arg− K+, pKD1+) (4) and MW103–1C (Mat a, uraA lysA, K+, pKD1+) (10). K. lactis were grown at 26°C either in yeast extract/peptone/dextrose or SD-u (synthetic uracil drop-out media) media [0.67% yeast nitrogen base without amino acids, 2% glucose, and 2% SCM-URA (commercial synthetic mix of amino acids and inositol) (Bufferad) (11)]. Escherichia coli DH5α and DH10B, from Life Technologies, were grown upon transformation in Luria–Bertani media with 100 μg/ml ampicillin.
Plasmids, Ligations, and Electroporation.
Plasmid EPGK31 (a gift from Hiroshi Fukuhara, Institute Curie) is a K. lactis/E. coli shuttle vector derived from Kep6 (12) containing the S. cerevisiae phosphoglycero kinase promoter and terminator regions flanking a BglII cloning site. Plasmids E1 and E4 were obtained by ligation in either orientation of the 57-bp BglII–BamHI fragment from pLitmus 28 (New England Biolabs) into the BglII site of pEPGK 31.
DNA ligations were performed with T4 DNA ligase (Life Technologies), and dsDNA probes were labeled with Oligolabeling kit (Pharmacia Biotech) and [α-32P]dCTP as indicated by the manufacturers. E. coli plasmid DNA was isolated either with Wizard Plus Miniprep kit (Promega) or Nucleobond AX (The Nest Group, Southport, MA). Plasmid DNA was isolated from yeast as described (13). All transformations were performed by electroporation as described (3).
Construction of a MDCK cDNA Library.
Total RNA was isolated from confluent MDCK cells by extraction with phenol and guanidine thiocyanate (Tri Reagent, Molecular Research Center, Cincinnati). Poly(A)+ mRNA was obtained from the former RNA by oligo(dT) cellulose affinity chromatography [Oligo(dT) prepacked columns, GIBCO/BRL]. First strand cDNA synthesis was accomplished by using the Super Script Choice System (Life Technologies). In brief, 5 μg of poly(A)+ RNA was transcribed into cDNA with 200 units of Superscript II avian myeloblastosis viral reverse transcriptase and 1 μg of oligo(dT) plus 5 ng of random hexameres as primers. After second strand synthesis with E. coli DNA polymerase I in combination with E. coli RNase H and E. coli DNA ligase, an Eco(Not) adapter was ligated to both ends of the cDNA.
The cDNA was size-selected by chromatography through Sephacryl S-500 HR and ligated into pE1 linearized with EcoRI and then transformed into E. coli ElectroMax DH10B by electroporation. Approximately 105 independent transformants were obtained with an average insert of 2.0 ± 0.6 kb. Plasmid DNA, extracted from 5 pools of ≈2 × 104 colonies each, was transformed into KL3 cells plated on uracil-free media. Approximately 5 × 104 colonies were collected from each pool of DNA (≈2 per original E. coli colony because of the nondirectionality of the cDNA cloning) and stored until further analysis in yeast extract/peptone/dextrose medium with 15% glycerol at −70°C).
Cell Surface Labeling of K. lactis with GS II-FITC and Fluorescent-Activated Cell Sorting.
K. lactis cells were washed with labeling buffer (50 mM sodium phosphate/150 mM NaCl/1 mM CaCl2/0.5 mM MgCl2/0.1 mM MnCl2, pH 7.0). Then, 1.5 OD600 of cells were incubated with 20 μl of GSII-FITC lectin (1 mg/ml) and 180 μl of labeling buffer for 2 h at 23°C vortexing every 15 min. After the incubation, 2 ml of ice-cold labeling buffer was added and the cells were pelleted by centrifugation at 4°C. Two more washings were performed with 3 ml of buffer in a similar manner, and finally the cells were resuspended in 1 ml of labeling buffer and kept on ice until passage through the cell sorter.
Cells were run through either a FACStar Plus or a FACS Vantage (Becton Dickinson) as described before (3). When individual clones were studied, a FACScan (Becton Dickinson) was used.
Sequencing and Data Analysis.
A 2-kb SalI fragment containing the entire cDNA insert of the UDP-GlcNAc transporter from p#37 clone was subcloned into pBluescript II SK(+) (Stratagene). DNA sequencing and primer synthesis were performed by an automated Applied Biosystems DNA sequencer with dye primers by using the dideoxy chain termination method at the Molecular Genetics Facility of the University of Georgia, Athens. Sequences were assembled and analyzed by using the DNAstar program (DNAstar, Madison, WI), and database searches were made through the NCBI Blast server (National Center for Biotechnology Information, Bethesda, MD). Reverse transcriptase PCR was performed by using the Superscript One-Step RT-PCR System (Life Technologies) and primers F238 (5′-CCTCACAAATTAGACCCAGAAC) and N253 (5′-TCTTCTACAGCAGTGGTTGTTG).
Subcellular Fractionation and Assays for Nucleotide Sugar Translocation and GDPase.
Golgi-enriched membranes from K. lactis were obtained as described (4). Latency of GDPase (14) was at least 94%, demonstrating that the vesicles were intact and of the same orientation as in vivo.
Translocation assays were performed as described (15) in 1 ml of 10 mM Tris⋅HCl buffer (pH 7.4) containing 0.25 M sucrose, 0.15 M KCl, 5 mM MgCl2, and 1 mM CaCl2. Incubations contained 0.4–0.9 mg of protein and 7 × 105 cpm of UDP-[3H]GlcNAc or GDP-[3H]mannose and were done for 5 min at 30°C. Total radioactive solutes within the vesicle pellet were determined and calculated as described (15). The volume outside vesicles in the pellet was 3.6 μl/mg using [3H]acetate as nonpenetrator (15).
RESULTS
We previously characterized the biochemical defect of a K. lactis mutant lacking terminal N-acetylglucosamine in its mannan chains (5) as a deficiency of transport of UDP-GlcNAc into the Golgi apparatus (4). In the Golgi lumen, this nucleotide sugar is a substrate for the α1–2 N-acetylglucosaminyltransferase that adds one terminal N-acetylglucosamine per repetitive mannan subunit. This mutant recently was used by us to clone the homologous Golgi membrane UDP-GlcNAc transporter gene by complementation (6). For this, we took advantage of the differences in binding at the cell surface of the GSII lectin between wild type and the above mutant because terminal N-acetylglucosamine specifically binds to the above fluorescent lectin whereas the mutant does not.
Our strategy to isolate the MDCK cDNA encoding the Golgi UDP-GlcNAc transporter relied on the isolation of those K. lactis mnn2–2 mutants that had acquired wild-type binding of GSII-FITC lectin and had appeared on the high fluorescence window of the FACS after transformation with a library. The library was prepared cloning the MDCK cDNA behind the strong yeast constitutive PGK promoter on a pKD1-derived shuttle vector, pE4. Plasmid DNA, obtained from 105 independent E. coli transformants, was used to generate 2.5 × 105 independent K. lactis mnn2–2 transformants, which were screened for the appearance of the N-acetylglucosamine epitope at the cell surface by GSII-FITC lectin binding and FACS. The same approach was used successfully to clone the K. lactis Golgi UDP-GlcNAc transporter (6).
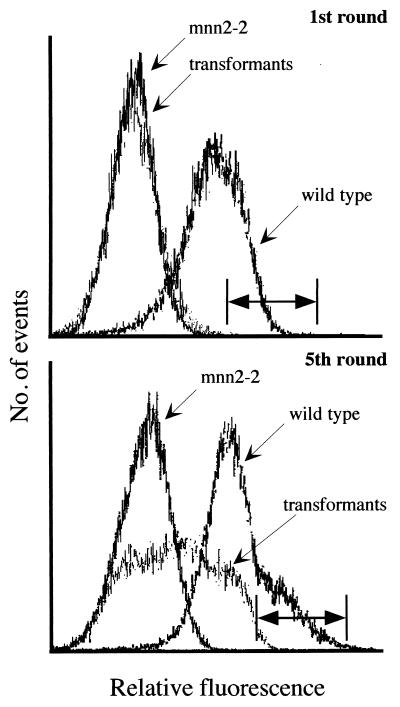
The fraction of the total cells showing wild-type fluorescence in the 0.1 percentile higher range was collected. These cells were grown overnight in uracil-free media and stored frozen until further analysis. The high fluorescent cells of the first round of sorting were pooled, subdivided in aliquots, labeled with GSII-FITC, and subjected to a total of five sorting cycles. After the latter sorting cycle, a significant increase in the population of cells binding to GSII-FITC like wild type could be seen (Fig. 1).
Figure 1.
K. lactis cell surface labeling and separation by FACS. (Upper) Golgi UDP-GlcNAc transporter-deficient K. lactis mutants (KL3; mnn2–2) were transformed with a MDCK cell cDNA library and screened by FACS. Terminal N-acetylglucosamine of wild-type K. lactis (MW103–1C) glycoproteins bind GSII lectin. These cells display higher fluorescence intensity than the UDP-GlcNAc Golgi transporter-deficient mutant. Transformed cells, after one cycle of cell sorting, show a fluorescence profile resembling the transporter-deficient mutant. The horizontal bar indicates the 0.1 percentile of high fluorescence of the transformed cells; cells whose fluorescence spectrum falls in this region were selected for subsequent rounds of cell sorting. (Lower) After five cycles of sorting and selection of the high fluorescence cells, a significant shift in the fluorescence profile of the transformants can be seen into the region of the wild-type cells. Cells whose fluorescence fell in this region, indicated by the horizontal bar, were selected for further analysis of individual clones.
Following the above fifth sorting cycle, cells in the 0.1 percentile of high fluorescence were plated onto uracil-free medium and single colonies were re-examined for the appearance of N-acetylglucosamine at the cell surface by their ability to bind the above lectin. Several of the clones showing wild-type GSII-FITC binding were isolated, although there was always a fraction, <30% of the total population, which showed fluorescence binding in the mutant range. Plasmids were recovered from these K. lactis mnn2–2 transformants, and cDNA inserts were analyzed. The smallest (2 kb) cDNA, which was shown by Southern hybridization to be contained in all the other larger inserts, was chosen for further characterization.
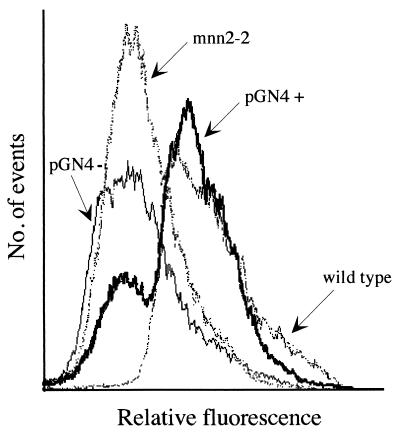
As a control that the phenotypic correction depended on the expression of the protein encoded by this cDNA, a SalI fragment, containing the entire insert, was ligated nondirectionally into the XhoI site of pE 4, the vector in which the library was made. This yielded a cDNA in both orientations with respect to the phosphoglycero kinase promoter with only the one in the positive orientation causing phenotypic correction (Fig. 2, Lower). The specificity of the above phenotypic correction was supported further by transforming the above KL3 mutant with plasmids pGal+ and pGal− containing the MDCK Golgi UDP-galactose transporter in both orientations with respect to the promoter (E.G., unpublished work). None of these plasmids caused phenotypic correction as determined by mutant-level of GSII binding (not shown).
Figure 2.
The cDNA in p37 restores wild-type GSII phenotype of the UDP-GlcNAc Golgi transporter mutant. The cDNA in the selected p37 clone was extracted and religated, in either orientation, in the expression vector pE4 under the control of the PGK constitutive promoter. Upon transformation into the Golgi UDP-GlcNAc transporter-deficient mutant, the plasmid containing the cDNA in the plus orientation with respect to the promoter (PGN4+) restored the fluorescence profile resembling that of wild-type cells. Cells transformed with the plasmid in the minus orientation (PGN4-) showed no phenotypic correction.
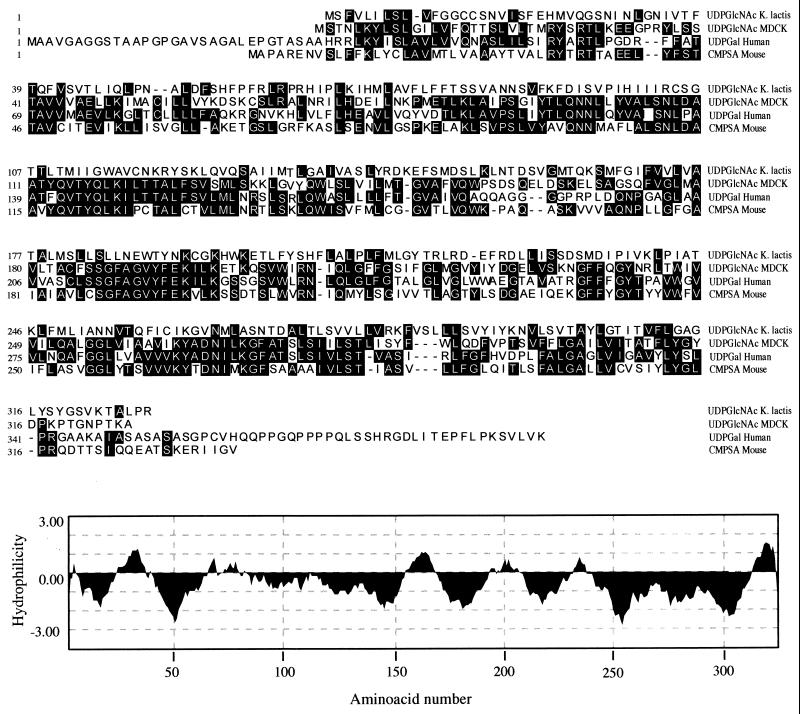
The isolated cDNA, which corrects the mutant phenotype, contains 1,960 bp and has an ORF of 326 aa encoding a very hydrophobic multitransmembrane-spanning protein of 36 kDa (Fig. 3). This protein has in its amino terminus an apparent noncleavable endoplasmic reticulum translocation signal. Amino acid sequence comparison with nucleotide sugar transporters of known functions reveals a surprisingly low (22%) identity with the UDP-GlcNAc transporter of K. lactis consistent with our lack of positive signals in Northern analyses (Fig. 3). The MDCK Golgi UDP-GlcNAc transporter, however, shows a significantly higher degree of amino acid sequence identity with the human UDP-galactose transporter (53%) and a 40% amino acid sequence identity with the human and murine CMP-sialic acid transporters (Fig. 3).
Figure 3.
(Upper) Amino acid alignment of the predicted sequences for the MDCK (AF057365) and K. lactis (U 48413) UDP-GlcNAc, human UDP-Gal (D84454), and mouse CMP-sialic acid (Q61420) transporters. (Lower) Hydrophilicity plot of the MDCK UDP-GlcNAc transporter using the Kyte–Doolittle algorithm (16) with a window of 17 aa.
To further demonstrate that the above transporter protein indeed is encoded by a MDCK mRNA, reverse transcriptase PCR was performed by using MDCK RNA preparation, not used in construction of the above library. F238 and N253 (see Materials and Methods) were used as primers. The expected 1.4-kb product was obtained and purified by agarose gel electrophoresis. Partial sequencing resulted in a sequence identical to that shown above.
It was important to determine whether the phenotypic correction of GSII lectin binding of the transformant correlated with UDP-GlcNAc transport into Golgi vesicles from these cells. A fraction enriched in Golgi vesicles from wild-type, mutant, and transformant was therefore tested for their ability to transport UDP-GlcNAc in vitro.
As shown in Table 1, the mnn 2–2 mutant shows no significant UDP-GlcNAc transport whereas wild-type and transformant-derived vesicles do. All vesicles have similar specific GDP-mannose transport activity, demonstrating that the UDP-GlcNAc transport defect correction of the transformant is specific and correlates with the observed in vivo binding with GSII lectin. The somewhat lower values of translocation of the transformant Golgi vesicles compared with those of the wild type are consistent with the observation that only a fraction of the transformed cells showed wild type cell levels of fluorescence.
Table 1.
Translocation of UDP-GlcNAc and GDP mannose into a Golgi-enriched vesicle fraction of K. lactis
| Solutes within vesicles, pmol/mg protein × 5 min
| |||
|---|---|---|---|
| Substrate | Wild type | Mutant mnn 2-2 | Mutant mnn 2-2 transformed with plasmid encoding the UDP-GlcNAc transporter |
| UDP-GlcNAc | 0.20 ± 0.05 | 0.02 ± 0.01 | 0.11 ± 0.01 |
| GDP-Man | 3.70 ± 0.38 | 2.41 ± 0.18 | 2.89 ± 0.03 |
UDP-[3H]GlcNAc (30,000 cpm/pmol) or GDP-[3H]mannose (4,000 cpm/pmol) were incubated with a P3 Golgi-enriched vesicle fraction (0.4–0.9 mg/protein) in 1 ml of buffer as described in Materials and Methods. Translocation was determined as described (15). Results are an average ± SD of three to five determinations.
DISCUSSION
Two important concepts have emerged from this study: (i) transport of UDP-N-acetylglucosamine can be mediated across the same Golgi membrane by two multitransmembrane spanning proteins that have a very different primary sequence; and (ii) the mammalian Golgi membrane UDP-N-acetylglucosamine transporter cDNA has all of the necessary information to express and target this protein in a functional manner to the Golgi apparatus of K. lactis so that it can provide the nucleotide sugar in the lumen of this organelle as substrate for the corresponding N-acetylglucosaminyltransferase to add N-acetylglucosamine to mannan chains, which then migrate to the cell surface and bind to GSII lectin as wild-type cells.
The limited amino acid sequence similarity or identity between the transporters for the same substrate in yeast and mammals is particularly striking in view of the high degree of sequence similarity between the transporters for CMP-sialic acid from murines, UDP-galactose from human, and now UDP-GlcNAc from MDCK cells. These results illustrate that, for the time being, knowledge of primary sequences of Golgi nucleotide sugar transporters cannot allow us to predict the substrate specificity of these proteins and that a given transporter function can be effected by very different proteins.
When we began the above described studies, we did not know whether a mammalian Golgi transporter protein could be expressed, be targeted to the Golgi apparatus, and remain active in a yeast system. While these studies were ongoing, we demonstrated that we were able to express the murine CMP-sialic acid Golgi transporter in S. cerevisiae and that a portion of this transporter was targeted to Golgi vesicles in a functionally active form. The studies described here extend the studies with the heterologous functional expression of the CMP-sialic acid transporters by demonstrating that the heterologously expressed transporter can indeed make the nucleotide sugar available in vivo for endogenous glycosyltransferases to use as substrate for the addition of the corresponding sugar to endogenous acceptors and subsequent transport of these macromolecules to the cell surface.
The Golgi targeting signals for these transporters are unknown, but these studies have shown that proteins with different primary sequences can be targeted correctly. Whether general Golgi targeting signals for these multitransmembrane-spanning proteins will be as difficult to elucidate as those for type II membrane proteins remains to be seen (17).
We recently described that MDCK cells synthesize keratan sulfate, chondroitin sulfate, and heparan sulfate, in addition to glycoproteins and glycolipids (3). A ricin-resistant mutant, which transports only 5% of wild-type levels of UDP-galactose into the Golgi lumen, showed, surprisingly, differential synthesis of galactose-containing proteoglycans: keratan sulfate was not synthesized while chondroitin sulfate and heparan sulfate were at normal levels (3). This research has provided evidence in mammalian cells that nucleotide-sugar transporter-mediated substrate availability drastically can affect which Golgi macromolecules are synthesized. MDCK cells contain heparan sulfate and keratan sulfate as N-acetylglucosamine-containing macromolecules, in addition to glycoproteins (3). It will be interesting to see whether overexpression of this and other Golgi nucleotide sugar transporters, such as that of UDP-galactose in these cells, leads to differential synthesis of glycoconjugates and whether the same can be observed under limited supply of UDP-N-acetylglucosamine. The latter perhaps will be accomplished by introducing an antisense RNA to the UDP-GlcNAc transporter into these cells or transfection with an inactive transporter that localizes to the Golgi apparatus.
Acknowledgments
We thank Glenn Paradis, Michael Jennings, and Michael Connolly (MIT Flow Cytometry Core Facility) for help with cell sorting, Dr. Hiroshi Fukuhara, Institute Curie, Orsay, France, for K. lactis strains, plasmids, and very helpful discussions, and Karen Welch for expert secretarial assistance. This work was supported by National Institutes of Health Grant GM 34396.
ABBREVIATIONS
- PGK
phosphoglycero kinase
- GSII
Griffonia simplicifolia II
- FACS
fluorescent-activated cell sorter
- FITC
fluorescein isothiocyanate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF057365).
References
- 1.Abeijon C, Mandon E C, Hirschberg C B. Trends Biochem Sci. 1997;22:203–207. doi: 10.1016/s0968-0004(97)01053-0. [DOI] [PubMed] [Google Scholar]
- 2.Abeijon C, Yanagisawa K, Mandon E C, Hausler A, Moremen K, Hirschberg C B, Robbins P W. J Cell Biol. 1993;122:307–327. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toma L, Pinhal M A S, Dietrich C P, Nader H B, Hirschberg C B. J Biol Chem. 1996;271:3897–3901. doi: 10.1074/jbc.271.7.3897. [DOI] [PubMed] [Google Scholar]
- 4.Abeijon C, Mandon E C, Robbins P W, Hirschberg J Biol Chem. 1996;271:8851–8856. doi: 10.1074/jbc.271.15.8851. [DOI] [PubMed] [Google Scholar]
- 5.Douglas R K, Ballou C E. Biochemistry. 1982;21:1561–1570. doi: 10.1021/bi00536a015. [DOI] [PubMed] [Google Scholar]
- 6.Abeijon C, Robbins P W, Hirschberg C B. Proc Natl Acad Sci USA. 1996;93:5963–5968. doi: 10.1073/pnas.93.12.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miura N, Ishida N, Hoshino M, Yamauchi M, Hara T, Ayusawa D, Kawakita M. J Biochem. 1996;120:236–241. doi: 10.1093/oxfordjournals.jbchem.a021404. [DOI] [PubMed] [Google Scholar]
- 8.Capasso J M, Hirschberg C B. Biochem Biophys Acta. 1984;777:133–139. doi: 10.1016/0005-2736(84)90505-4. [DOI] [PubMed] [Google Scholar]
- 9.Brandli A W, Hansson G C, Rodriguez-Boulan E, Simons K. J Biol Chem. 1988;263:16283–16290. [PubMed] [Google Scholar]
- 10.Chen X J, Wesolowski-Louvel M, Tanguy-Rougeau C, Fukuhara H. Biochemie. 1991;73:1195–1203. doi: 10.1016/0300-9084(91)90004-k. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser C, Michaelis M, Mitchel A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 12.Chen X J, Wesolowski-Louvel M, Tanguy-Rougeau C, Bianchi M M, Fabiani L, Saliola M, Falcone C, Frontali L, Fukuhara H. J Basic Microbiol. 1988;28:211–220. doi: 10.1002/jobm.3620280402. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman C S. Curr Protocols Mol Biol. 1997;13:11.1. [Google Scholar]
- 14.Abeijon C, Orlean P, Robbins P W, Hirschberg C B. Proc Natl Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez M, Hirschberg C B. Methods Enzymol. 1987;138:709–715. doi: 10.1016/0076-6879(87)38061-9. [DOI] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Colley K J. Glycobiology. 1997;7:1–13. doi: 10.1093/glycob/7.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]