Abstract
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. The N-end rule pathway is one proteolytic pathway of the ubiquitin system. The recognition component of this pathway, called N-recognin or E3, binds to a destabilizing N-terminal residue of a substrate protein and participates in the formation of a substrate-linked multiubiquitin chain. We report the cloning of the mouse and human Ubr1 cDNAs and genes that encode a mammalian N-recognin called E3α. Mouse UBR1p (E3α) is a 1,757-residue (200-kDa) protein that contains regions of sequence similarity to the 225-kDa Ubr1p of the yeast Saccharomyces cerevisiae. Mouse and human UBR1p have apparent homologs in other eukaryotes as well, thus defining a distinct family of proteins, the UBR family. The residues essential for substrate recognition by the yeast Ubr1p are conserved in the mouse UBR1p. The regions of similarity among the UBR family members include a putative zinc finger and RING-H2 finger, another zinc-binding domain. Ubr1 is located in the middle of mouse chromosome 2 and in the syntenic 15q15-q21.1 region of human chromosome 15. Mouse Ubr1 spans ≈120 kilobases of genomic DNA and contains ≈50 exons. Ubr1 is ubiquitously expressed in adults, with skeletal muscle and heart being the sites of highest expression. In mouse embryos, the Ubr1 expression is highest in the branchial arches and in the tail and limb buds. The cloning of Ubr1 makes possible the construction of Ubr1-lacking mouse strains, a prerequisite for the functional understanding of the mammalian N-end rule pathway.
Keywords: ubiquitin/proteolysis/E3/N-recognin/Ubr1
A number of regulatory circuits involve metabolically unstable proteins. Short in vivo half-lives are also characteristic of damaged or otherwise abnormal proteins (1–4). Features of proteins that confer metabolic instability are called degradation signals, or degrons. The essential component of one degradation signal, called the N-degron, is a destabilizing N-terminal residue of a protein (5, 6). The set of amino acid residues that are destabilizing in a given cell type yields a rule, called the N-end rule, which relates the in vivo half-life of a protein to the identity of its N-terminal residue. Similar, but distinct, versions of the N-end rule pathway are present in all organisms examined, from mammals to fungi and bacteria (6–8).
In eukaryotes, the N-degron comprises two determinants: a destabilizing N-terminal residue and an internal lysine or lysines (8). The Lys residue is the site of formation of a multiubiquitin chain (9). The N-end rule pathway is thus one pathway of the ubiquitin (Ub) system. Ub is a 76-residue protein whose covalent conjugation to other proteins plays a role in a multitude of processes, including cell growth, division, differentiation, and responses to stress (1, 3, 4, 10). In most of these processes, Ub acts through routes that involve the degradation of Ub-protein conjugates by the 26S proteasome, an ATP-dependent multisubunit protease (11).
The N-end rule is organized hierarchically. In the yeast Saccharomyces cerevisiae, Asn and Gln are tertiary destabilizing N-terminal residues in that they function through their enzymatic deamidation into the secondary destabilizing N-terminal residues Asp and Glu (12). The destabilizing activity of N-terminal Asp and Glu requires their enzymatic conjugation to Arg, one of the primary destabilizing residues (6). The primary destabilizing N-terminal residues are bound directly by the UBR1-encoded N-recognin (also called E3), the recognition component of the N-end rule pathway (13). In S. cerevisiae, N-recognin is a 225-kDa protein that binds to potential N-end rule substrates through their primary destabilizing N-terminal residues—Phe, Leu, Trp, Tyr, Ile, Arg, Lys, and His. N-recognin has at least two substrate-binding sites. The type 1 site is specific for the basic N-terminal residues Arg, Lys, and His. The type 2 site is specific for the bulky hydrophobic N-terminal residues Phe, Leu, Trp, Tyr, and Ile (6).
The known functions of the N-end rule pathway include the control of peptide import in S. cerevisiae (through degradation of Cup9p, a transcriptional repressor of the peptide transporter Ptr2p); a role in controlling the Sln1p-dependent phosphorylation cascade that mediates osmoregulation in S. cerevisiae; the degradation of Gpa1p, a Gα protein of S. cerevisiae; and the degradation of alphaviral RNA polymerases in virus-infected metazoan cells (6, 14).
The mammalian counterpart of the yeast UBR1-encoded N-recognin (E3) was characterized biochemically in extracts from rabbit reticulocytes (15–17). Rabbit E3α was shown to be specifically required for the Ub-dependent degradation of proteins bearing either type 1 (basic) or type 2 (bulky hydrophobic) destabilizing N-terminal residues (7, 15, 16).
We began dissection of the mouse N-end rule pathway by isolating the Ntan1 gene, which encodes the asparagine-specific N-terminal amidase (18, 19), a component of the mammalian N-end rule pathway, and by constructing mouse strains that lack Ntan1 (Y.T.K. and A.V., unpublished data). Herein, we describe the cloning and characterization of the mouse and human cDNAs and genes‡‡ that encode UBR1p (E3α), a homolog of yeast Ubr1p and the main recognition component of the N-end rule pathway.
MATERIALS AND METHODS
Isolation and Partial Sequencing of Mammalian E3α (UBR1p).
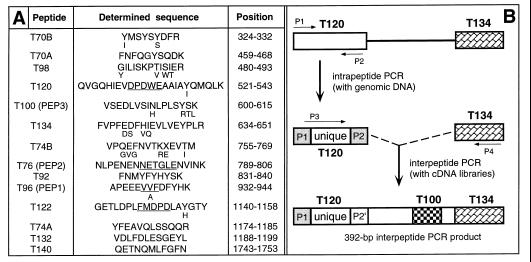
Rabbit E3α was purified from reticulocyte extracts by using affinity chromatography with immobilized protein substrates of UBR1p and elution with dipeptides bearing destabilizing N-terminal residues (16). The resulting preparation was fractionated by SDS/PAGE. The band of ≈180-kDa E3α was excised and subjected to digestion with trypsin. Amino acid sequences were determined for 14 peptides of rabbit UBR1p (Fig. 1A) by using standard methods (20).
Figure 1.
Peptides of rabbit UBR1p (E3α) and isolation of the mouse Ubr1 cDNA. (A) Amino acid sequences of tryptic peptides of the purified rabbit UBR1p (see Materials and Methods). The alternative sets of peptide names, T-based and PEP1-PEP3 (in parentheses), refer to two different preparations of E3α. The sequences of T120, T76, T96, and T122 that were encoded by DNA sequences identified through intrapeptide PCR are underlined. Residues deduced from the mouse Ubr1 cDNA that differed from those inferred through peptide sequencing are indicated in a smaller font. The peptides’ positions in the deduced sequence of mouse UBR1p are indicated. (B) The intrapeptide/interpeptide-PCR cloning strategy. The products of the initial intrapeptide PCR, derived from rabbit genomic DNA, were used to carry out interpeptide PCRwith a rabbit liver cDNA library (CLONTECH). The resulting 392-bp fragment of the rabbit Ubr1 cDNA was used to isolate, using PCR and a λgt11 mouse liver cDNA library, the corresponding 392-bp mouse Ubr1 cDNA fragment. This fragment then was used to screen the same cDNA library, yielding a 2.4-kb fragment of the mouse Ubr1 cDNA that encoded several of the peptide-derived sequences of the rabbit UBR1p. The encoded sequence was also significantly similar to that of the N-terminal region of S. cerevisiae Ubr1p (13) and contained the putative start (ATG) codon of the mouse Ubr1 ORF. To isolate the rest of the 5′ region of the Ubr1 cDNA, 5′-rapid amplification of cDNA ends (RACE)–PCR (20) was performed with poly(A)+ RNA from mouse L cells and a primer from the 2.4-kb DNA fragment. 3′-RACE–PCR (20) was used to amplify a downstream region of Ubr1 cDNA. The resulting DNA fragment (nucleotides 2,470–3,467) then was used to screen a λgt10 mouse cDNA library from MEL-C19 cells. Five overlapping cDNA isolates (MR16, MR17, MR19, MR20, and MR23) that together spanned the entire Ubr1 cDNA were mapped and subcloned into Bluescript II SK+ (Stratagene), yielding the plasmid MR26, which contained the entire ORF of Ubr1. The ORF region of Ubr1 cDNA was sequenced on both strands at least twice, using independently derived cDNA clones.
Isolation of the Full-Length Mouse Ubr1 cDNA.
A strategy that included the intrapeptide-interpeptide PCR (21) was used (see the legend to Fig. 1).
Isolation of a Partial Human UBR1 cDNA.
Poly(A)+ RNA from human 293 cells was subjected to reverse transcription–PCR, using sets of primers corresponding to sequences of the mouse Ubr1 cDNA. One of the reactions yielded a 1.0-kilobase (kb) fragment that encompassed a region of the human UBR1 cDNA (Fig. 2).
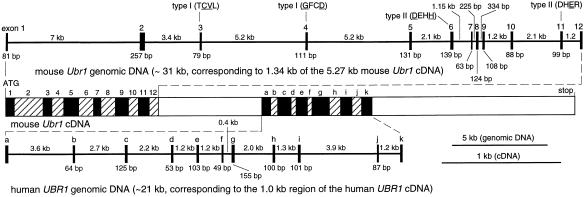
Figure 2.
The mouse and human Ubr1 cDNAs and genes. Thick horizontal lines represent genomic DNA. The upper one is a ≈31-kb fragment of the mouse Ubr1 gene that corresponds to a 1.34-kb region of mouse Ubr1 cDNA (nucleotides 115–1,454). Vertical rectangles represent exons. Their lengths, and the lengths of the introns, are indicated, respectively, below and above the horizontal line. In a composite diagram of the Ubr1 cDNA, the exons are depicted as alternatively shaded rectangles. For exon 1, only its translated region is indicated. Shown belowthe cDNA diagram is a ≈21-kb fragment of the human UBR1 gene, corresponding to 1.0 kb of the indicated region of the human UBR1 cDNA (nucleotides 2,218–3,227 of the mouse Ubr1 cDNA sequence). The mouse and human Ubr1 exons are denoted, respectively, by numbers and letters. Also indicated are the exon locations of some of the type 1 and type 2 substrate-binding sites of N-recognin (the essential amino acid residues are underlined) (A. Webster, M. Ghislain, and A.V., unpublished data; see the main text). Not shown are the 114-bp 5′-untranslated region (UTR) and the 1,010 bp 3′-UTR of the mouse Ubr1 cDNA. To isolate mouse Ubr1, a library of mouse genomic DNA fragments in a BAC vector (see Materials and Methods) was screened with a fragment of the mouse Ubr1 cDNA (nucleotides 105–1,333) as a probe, yielding seven BAC clones, of which BAC3 and BAC4 contained the entire Ubr1 gene. The exon/intron organization of the first 31 kb (≈1/4) of the mouse Ubr1 gene was determined by using exon-specific PCR primers to produce ≈40 genomic DNA fragments of the BAC3 insert that ranged in size from 1.3 to 18 kb. Regions encompassing the exon/intron junctions then were sequenced by using intron-specific primers. Fragments of the human genomic UBR1 DNA were isolated by using primers derived from the 1.0-kb fragment of the human UBR1 cDNA, the Expand High Fidelity PCR System (Roche Molecular Biochemicals, Indianapolis, IN), and genomic DNA from human 293 cells. The resulting four fragments were subcloned into pCR2.1 (Invitrogen), yielding the plasmids HR8, HR6–4, HR2–25, and HR7–2, whose partially overlapping inserts encompassed ≈21 kb of the human UBR1 gene. Partial sequencing of the mouse and human genomic Ubr1 fragments (≈20 kb of sequenced DNA) included all of the exon/intron junctions in these regions of Ubr1.
Mouse and Human Genomic Ubr1 Fragments.
A library of mouse genomic DNA fragments (strain SvJ) in bacterial artificial chromosome (BAC) (22) vector (Genome Systems, St. Louis) was used, as described in the legend to Fig. 2.
Northern, Southern, and Whole-Mount in Situ Hybridizations.
Mouse and human multiple-tissue Northern blots (CLONTECH), and either mouse or human Ubr1 cDNA fragments labeled with 32P were used (20). Southern hybridizations were carried out by using standard techniques (20). Mouse embryos were staged, fixed, and processed for in situ hybridization as described (23). For sectioning, the stained embryos were embedded in OCT medium (Sakura Finetek, Torrance, CA). A 1.2-kb Ubr1 cDNA fragment (nucleotides 3,150–3,355) was used as a template for synthesizing antisense- or sense-strand RNA probes labeled with digoxigenin (23).
Chromosome Mapping of the Mouse and Human Ubr1.
The mapping of mouse Ubr1 was carried out by using the interspecific backcross analysis (24), essentially as described (18). Human UBR1 was mapped by using fluorescence in situ hybridization (FISH) with mitotic chromosomes from human lymphocytes (25). The probe was a mixture of the HR8, HR6–4, HR2–25, and HR7–2 plasmids, labeled with biotin using biotinylated dATP and the BioNick labeling kit (Life Technologies, Grand Island, NY), and detected by using fluorescein isothiocyanate-avidin.
RESULTS AND DISCUSSION
Isolation of the Mouse Ubr1 cDNA.
Tryptic peptides of the purified rabbit E3α (UBR1p) (16) were isolated and sequenced (see Materials and Methods), yielding 14 short regions of E3α (Fig. 1A). These regions lacked significant similarities to the deduced sequence of S. cerevisiae Ubr1p (13). We used intrapeptide PCR (21) to identify a unique (nondegenerate) sequence of the rabbit Ubr1 cDNA. This method allows amplification of a short unique DNA sequence by using two degenerate PCR primers (derived by reverse translation) that flank this sequence and correspond to the outermost regions of a single peptide (Fig. 1B). Several intrapeptide nucleotide sequences were obtained this way (Fig. 1). These sequences, together with those of the original degenerate primers, then were used to amplify a 392-bp fragment of the rabbit Ubr1 cDNA, using interpeptide PCR (Fig. 1). This fragment encoded peptides T120 and T134 at either end and peptide T100 in the middle (Fig. 1B). A homologous 392-bp fragment of the mouse Ubr1 cDNA then was amplified by using the same method (Fig. 1B). The rabbit and mouse 392-bp Ubr1 cDNA fragments were 88% and 89% identical at the nucleotide and amino acid sequence levels, respectively, but lacked significant similarities to S. cerevisiae UBR1 (data not shown).
The 392-bp mouse Ubr1 cDNA fragment then was used, in conjunction with standard cDNA library screening and rapid amplification of cDNA ends–PCR (20), to isolate multiple Ubr1 cDNA fragments, and to assemble them into a 5,271-bp ORF encoding a 1,757-residue protein (pI of 6.0), whose size, 200 kDa, was close to the estimated size of the isolated rabbit UBR1p (E3α), ≈180 kDa (16) (Figs. 2 and 3). The inferred ATG start codon (Fig. 2), within the sequence CTTAAGATGGCG, is preceded by two in-frame stop codons, at positions −48 and −93, and is located in a favorable Kozak context (26), with A and G at positions −3 and +4, respectively. There are two more ATGs, five and 11 codons downstream of the inferred one. These alternative start codons are in a favorable Kozak context as well.
Figure 3.
Comparison of the deduced amino acid sequence of mouse UBR1p (Mm-UBR1) with those of C. elegans UBR1p (Ce-UBR1), S. cerevisiae Ubr1p (Sc-UBR1), and K. lactis Ubr1p (Kl-UBR1). White-on-black and gray shadings highlight, respectively, identical and similar residues. The residues of UBR proteins that are identical to those of S. cerevisiae Ubr1p are denoted by double dots, at positions where the identity involves just one non-cerevisiae protein. Also indicated are the regions of significant similarity among the four proteins. K. lactis UBR1 was cloned through its crosshybridization to S. cerevisiae UBR1 (P. Waller and A.V., unpublished data).
Cloning and Partial Characterization of the Mouse and Human Ubr1 Genes.
A fragment of the mouse Ubr1 cDNA was used to isolate a ≈120-kb mouse Ubr1 genomic DNA clone, carried in a BAC vector (22). We determined the exon/intron organization and restriction map of the ≈31-kb region of Ubr1 that corresponded to the 1,340-bp 5′-region of the mouse Ubr1 cDNA (nucleotides 105–1,333) (Fig. 2). The lengths of the 12 exons in this region of mouse Ubr1 range from 63 to 257 bp (Fig. 2).
The nucleotide and deduced amino acid sequences of the 1.0-kb human UBR1 cDNA fragment (see Materials and Methods), located approximately in the middle of UBR1 cDNA (nucleotides 2,218–3,227 of the mouse Ubr1 cDNA) (Fig. 2), were, respectively, 91% and 94% identical to the corresponding mouse Ubr1 cDNA and UBR1p sequences. Overlapping genomic DNA fragments of human UBR1 that, together, encompassed a ≈21-kb region of the human UBR1 gene and corresponded to the 1.0-kb fragment of the human UBR1 cDNA (Fig. 2), were isolated from human DNA by using cDNA-derived primers and PCR. Partial sequencing showed that this ≈21-kb region of human UBR1 contained 11 exons whose length ranged from 49 to 155 bp, a distribution of exon lengths similar to that in a different region of mouse Ubr1 (Fig. 2). All of the sequenced exon/intron junctions (≈23 exons), which encompassed a ≈52-kb region of the mouse and human Ubr1, contained the consensus GT and AG dinucleotides characteristic of the mammalian nuclear pre-mRNA splice sites (data not shown) (20). Extrapolating from these data on the mouse and human Ubr1 genes and the corresponding regions of their cDNAs (Fig. 2), a mammalian Ubr1 gene is expected to be ≈120 kb long and to contain ≈50 exons.
The Mouse UBR1p Protein and its Homologs.
The low overall sequence similarity of mouse UBR1p (E3α) to Ubr1p of either S. cerevisiae [22% identity (id.), 48% similarity (si.)] or another budding yeast, Kluyveromyces lactis (21% id., 48% si.), belied the presence of five regions, denoted I–V, which were significantly similar between the mouse and yeast versions of UBR1p (Figs. 3 and 4). By contrast, the Ntan1-encoded asparagine-specific N-terminal amidase, the most upstream component of the mouse N-end rule pathway, lacks sequence similarities to its S. cerevisiae counterpart Nta1p (12, 18). Database searches identified other likely homologs of mouse UBR1p, in particular the 1,927-residue protein of the nematode Caenorhabditis elegans (GenBank accession no. U88308) (32% id., 53% si.; termed Ce-Ubr1); the 1,872-residue S. cerevisiae protein (GenBank accession no. Z73196) (21% id., 47% si.; termed Sc-UBR2; ref. 4); the 2,168-residue C. elegans protein (GenBank accession no. U40029) (21% id., 45% si.; termed Ce-Ubr2); and the 794-residue CER3p protein of the plant Arabidopsis thaliana (GenBank accession no. X95962) (26% id., 49% si.). CER3p is involved in wax biosynthesis in A. thaliana (27). In addition, a 147-residue sequence of the yeast Candida albicans (http://alces.med.umn.edu/bin/genelist?LUBR1) was similar to the N-terminal region of mouse UBR1p (Fig. 4).
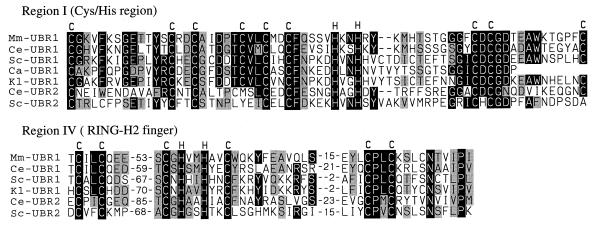
Figure 4.
Two Cys/His domains of the UBR protein family. Comparison of the putative zinc finger (region I) and RING-H2 finger (region IV) with the corresponding sequences from the other species in Fig. 3, and also with C. albicans Ubr1p (Ca-UBR1), C. elegans UBR2p (Ce-UBR2), and S. cerevisiae Ubr2p (Sc-UBR2). Numbers indicate the lengths of gaps. The conserved Cys and His residues are indicated.
The presence of high-similarity regions I–V among these deduced sequences (Figs. 3 and 4) suggested the existence of a distinct protein family, termed UBR. The 66-residue region I, near the N terminus of UBR1p, is a particularly clear UBR family-identifying region (e.g., 61% id., 75% si. between mouse and C. elegans UBR1p) (Figs. 3 and 4).
Recent genetic analyses of S. cerevisiae Ubr1p (N-recognin) have shown that the regions I–III contain residues essential for the recognition of N-end rule substrates by Ubr1p. In particular, Cys-145, Val-146, Gly-173, and Asp-176 of region I were identified as essential residues of the type 1 binding site of S. cerevisiae Ubr1p (A. Webster, M. Ghislain, and A.V., unpublished data). All four of these residues were conserved between the yeast, mouse, and C. elegans UBR1p (Figs. 3 and 4). Region I is present in all of the known UBR family members except CER3p of A. thaliana, which contains only regions IV and V (Fig. 4). Region I encompasses a Cys/His-rich domain, Cys-X12-Cys-X2-Cys-X5-Cys-X2-Cys-X2-Cys-X5-His-X2-His-X(12–14)-Cys-X1-Cys-X11-Cys (Figs. 3 and 4), which is distinct from the known consensus sequences of zinc fingers and other Cys/His-motifs. Residues Asp-318, His-321, and Glu-560 of S. cerevisiae Ubr1p, which have been identified as essential for the type 2 binding site of this N-recognin (A. Webster, M. Ghislain, and A.V., unpublished data), were found to be retained in region II (Asp-318 and His-321) and region III (Glu-560) of the mouse and C. elegans UBR1p (Fig. 3).
Region IV contains another Cys/His-rich domain of UBR1p, Cys-X2-Cys-loop 1-Cys-X1-His-X2-His-X2-Cys-loop 2-Cys-X2-Cys (Figs. 3 and 4), which is present in all of the UBR family members, and fits the consensus sequence of the RING-H2 finger, a subfamily of the previously defined RING motif (28). At least some of the RING-H2 sequences are sites of specific protein–protein interactions (28). Apc11p, a subunit of the Ub-protein ligase complex called the cyclosome (2) or the anaphase promoting complex, also contains a RING-H2 finger (29).
Another area of similarity (24–50% id., 46–70% si.) among the UBR family members is region V (Fig. 3 and data not shown). This region, 115 residues long in mouse UBR1p, near the protein’s C terminus, is particularly similar between mouse and C. elegans UBR1p (50% id., 70% si.) (Fig. 3). Region V is located 4–14 residues from the UBR proteins’ C termini, the exceptions being the S. cerevisiae and K. lactis Ubr1p, which bear, respectively, 132- and 159-residue tails of unknown function that are rich in the acidic Asp/Glu residues (36% and 33%) (Fig. 3). No significant similarities could be detected between mammalian UBR1p and other E3s (recognins) of the metazoan Ub system, including E6AP (30) and subunits of the cyclosome/anaphase promoting complex, except for the presence of a RING-H2 finger domain in the latter (29). [Different E3 proteins of the Ub system recognize different degrons in protein substrates, thereby defining distinct Ub-dependent proteolytic pathways (1, 4).]
Expression of Mouse and Human Ubr1.
The 5′- and 3′-proximal mouse cDNA probes yielded similar results, detecting a single ≈8-kb transcript in several tissues (Fig. 5 Aa and Ab). In the testis, however, the ≈8-kb species of Ubr1 mRNA was a minor one, the major species being ≈6 kb (Fig. 5Aa). The levels of either mouse or human Ubr1 mRNA were highest in skeletal muscle and heart (Fig. 5A). The expression of mRNA encoding E214K, one of the mouse Ub-conjugating (E2) enzymes and a likely component of the mouse N-end rule pathway (6), was also highest in skeletal muscle and heart (18).
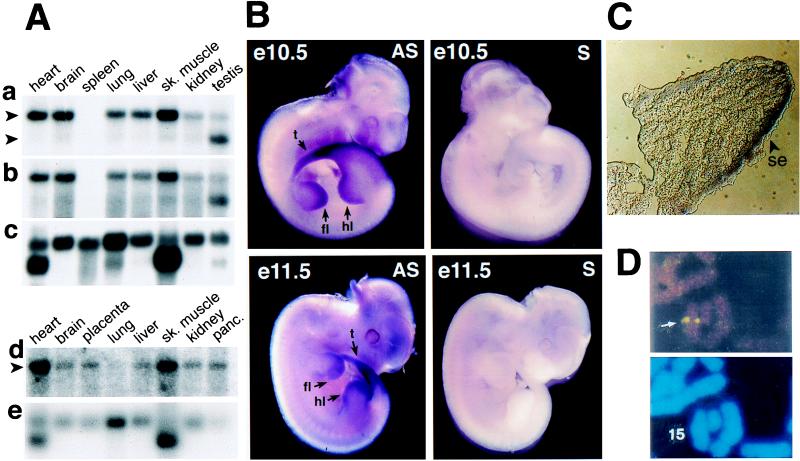
Figure 5.
Northern and in situ hybridizations with mouse and human Ubr1. (A) Membranes containing electrophoretically fractionated poly(A)+ mRNA from different mouse (a–c) or human (d and e) tissues were hybridized with either a 2-kb 5′-proximal (nucleotides 116–2,124) mouse Ubr1 cDNA fragment (a), its 0.64-kb 3′-proximal (nucleotides 4,749–5,388) fragment (b), a 1-kb human UBR1 cDNA fragment (d), or the human β-actin cDNA fragment (c and e). The upper arrows in a and d indicate the ≈8-kb Ubr1 transcript. The lower arrow in a indicates the ≈6-kb testis-specific Ubr1 transcript. In the RNA sample from mouse spleen, the Ubr1 transcript (but not the actin transcript) may have been degraded (a–c). (B) Expression of Ubr1 in e10.5 and e11.5 mouse embryos. Whole-mount in situ hybridization was carried out with either antisense (AS) or sense (S, negative control) Ubr1 cDNA probes (see Materials and Methods). The regions of high Ubr1 expression are indicated by arrows (t, tail; fl, forelimb buds; hl, hindlimb buds). The branchial arches, where Ubr1 is also highly expressed in e10.5 embryos (data not shown), are not visible in this e10.5 embryo. (C) Expression of Ubr1 in the surface ectoderm of limb buds. Shown is a transverse section of a forelimb bud of an e10.5 embryo (se, surface ectoderm). (D) FISH analysis of human UBR1. (Upper) An example of the UBR1-specific FISH signal (arrow). (Lower) The same mitotic spread stained with 4′-6-diamino-2-phenylindole (DAPI) to visualize the chromosomes (see also Fig. 6).
The distinct Ubr1 mRNA pattern in the testis (Fig. 5A) was reminiscent of the analogous expression pattern of Ntan1 mRNA, which encodes the Asn-specific N-terminal amidase, another component of the mammalian N-end rule pathway. Specifically, the size of the major species of Ntan1 mRNA was ≈1.4 kb in all of the examined mouse tissues except testis, where the major species was ≈1.1 kb (18). The ≈1.1-kb Ntan1 transcript recently was found to hybridize only to the 3′-half (exons 6–10 but not exons 1–5) of the Ntan1 ORF (Y.T.K. and A.V., unpublished data). The functional significance of the testis-specific Ubr1 and Ntan1 expression patterns remains to be understood.
We used whole-mount in situ hybridization to examine the expression of Ubr1 during embryogenesis. In e9.5 (9.5 days old) mouse embryos, the expression of Ubr1 was highest in the branchial arches and in the buds of forelimbs and the tail (data not shown). In e10.5 embryos, the expression of Ubr1 became high in the hindlimb buds as well (Fig. 5B). This pattern was maintained in the limb buds of e11.5 embryos (Fig. 5B). High expression of Ubr1 in the limb buds was confined predominantly to the surface ectoderm (Fig. 5C). This pattern of Ubr1 expression in embryos (Fig. 5 B and C) is similar, if not identical, to that of Ntan1, which encodes asparagine-specific N-terminal amidase (18) (Y.T.K. and A.V., unpublished data), consistent with UBR1p and NTAN1p being components of the same pathway.
The enhanced expression of Ubr1 in the embryonic limb buds (Fig. 5 B and C) is interesting in view of the conjecture that the N-end rule pathway might be required for limb regeneration in amphibians (31). The injection of dipeptides bearing destabilizing N-terminal residues into the stumps of amputated forelimbs of the newt was observed to delay limb regeneration, whereas the injection of dipeptides bearing stabilizing N-terminal residues had no effect (31). Rigorous tests of this and other suggested functions of the metazoan N-end rule pathway (6) will require mouse strains that lack Ubr1.
Chromosome Mapping of Mouse and Human Ubr1.
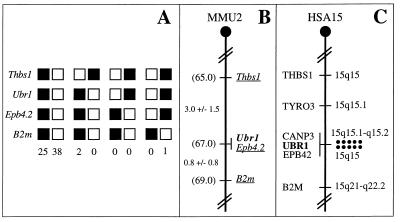
The chromosomal location of mouse Ubr1 was determined by interspecific backcross analysis, using DNA derived from matings of [(C57BL/6J × Mus spretus)F1 × C57BL/6J] mice (Fig. 6 A and B) (18, 24). Mouse Ubr1 is located in the central region of chromosome 2 and is linked to the Thbs1, Epb4.2, and B2m genes, the most likely gene order being centromere-Thbs1-Ubr1-Epb4.2-B2m (Fig. 6B and data not shown).
Figure 6.
Chromosomal locations of the mouse and human Ubr1 genes. (A) Mouse Ubr1 was mapped to the middle of mouse chromosome 2 by using interspecific (M. musculus-M. spretus) backcross analysis (18, 24). Shown are the segregation patterns of mouse Ubr1 and the flanking genes in 66 backcross animals that were typed for all loci. For individual pairs of loci, more than 66 animals were typed. Each column represents the chromosome identified in the backcross progeny that was inherited from the [M. musculus C57BL/6J × M. spretus] F1 parent. Filled and empty squares represent, respectively, C57BL/6J and M. spretus alleles. The numbers of offspring that inherited each type of chromosome 2 are listed below the columns. (B) A partial mouse chromosome 2 linkage map (MMU2), showing Ubr1 in relation to the linked genes Thbs1, Ebp4.2, and B2m, and also, on the left, the corresponding recombination distances between the loci, in centimorgans, and the map locations, in parentheses. (C) A partial human chromosome 15 linkage map (HSA15). Each dot on the right, in the 15q15-q21.1 region, corresponds to the actually observed UBR1-specific double-dot FISH signal detected on human chromosome 15 (see also Fig. 5D).
The chromosomal location of human UBR1 was determined by using FISH (25), with human UBR1 genomic DNA fragments as probes (Figs. 5D and 6C). This mapping placed UBR1 at the 15q15–15q21.1 region of the human chromosome 15, an area syntenic with the independently mapped position of mouse Ubr1 (Fig. 6). Ubr1 is located in the regions of human chromosome 15 and mouse chromosome 2 that appear to be devoid of the previously mapped but uncloned mutations. Mutations in the human gene CANP3, which encodes a subunit of calpain and is located very close, if not adjacent, to UBR1, have been shown to cause a myopathy called the limb-girdle muscular distrophy (32).
Concluding Remarks.
Isolation of the mouse and human Ubr1 cDNAs and genes (Figs. 2–6) should enable functional understanding of the mammalian N-end rule pathway, in part through the construction and analysis of mouse strains that lack Ubr1. Recent searches in GenBank identified several mouse and human sequences in expressed sequence tag databases that exhibited significant similarity to the C-terminal region of mouse UBR1p. The cloning and characterization of the corresponding cDNAs have shown that there exist at least two distinct mouse (and human) genes, termed Ubr2 and Ubr3, which encode proteins that are significantly similar to mouse UBR1p (Y.T.K. and A.V., unpublished data). Molecular and functional analyses of these Ubr1 homologs are under way.
Acknowledgments
We are grateful to A. Webster and M. Ghislain for permission to cite their unpublished data. We thank members of the Varshavsky lab, especially I. V. Davydov, for helpful discussions, and L. Peck, G. Turner, H. Rao, A. Kashina, and F. Du for comments on the manuscript. Y.T.K. thanks B. Yu for sharing his Northern hybridization data on human β-actin mRNA. We gratefully acknowledge the sequencing of K. lactis UBR1 by P. Waller. N.G.C. and N.A.J. thank D. J. Gilbert and D. B. Householder for excellent technical assistance. D.K.G. was a Scholar of the Leukemia Society of America. This study was supported by National Institutes of Health grants to A.V. (DK39520 and GM31530), V.A.F. (NS29542), and D.K.G. (GM45314), and by a grant to N.G.C. from the National Cancer Institute.
ABBREVIATIONS
- Ub
ubiquitin
- kb
kilobase
- id.
identity
- si.
similarity
- BAC
bacterial artificial chromosome
- FISH
fluorescence in situ hybridization
- en
embryonic day
Footnotes
Data deposition: Nucleotide sequences reported in this work have been deposited in the GenBank database [accession nos. AF061555 (mouse Ubr1 cDNA) and AF061556 (human UBR1 cDNA)].
The names of mouse genes are in italics, with the first letter uppercase. The names of human and S. cerevisiae genes are also in italics, all uppercase. If human and mouse genes are named in the same sentence, the mouse gene notation is used. The names of S. cerevisiae proteins are Roman, with the first letter uppercase and an extra lowercase “p” at the end. The names of the corresponding mouse and human proteins are the same, except that all letters but the last “p” are uppercase. The latter usage is a modification of the existing convention (33), to facilitate simultaneous discussions of yeast, mouse, and human proteins. In some citations, the abbreviated name of a species precedes the gene’s name.
References
- 1.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 3.Haas A J, Siepman T J. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 5.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 6.Varshavsky A. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 7.Gonda D K, Bachmair A, Wünning I, Tobias J W, Lane W S, Varshavsky A. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 8.Bachmair A, Varshavsky A. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 9.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 10.Pickart C M. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 11.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 12.Baker R T, Varshavsky A. J Biol Chem. 1995;270:12065–12074. doi: 10.1074/jbc.270.20.12065. [DOI] [PubMed] [Google Scholar]
- 13.Bartel B, Wünning I, Varshavsky A. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrd C, Turner G C, Varshavsky A. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiss Y, Kaim D, Hershko A. J Biol Chem. 1988;263:2693–269. [PubMed] [Google Scholar]
- 16.Reiss Y, Hershko A. J Biol Chem. 1990;265:3685–3690. [PubMed] [Google Scholar]
- 17.Hershko A, Ciechanover A. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 18.Grigoryev S, Stewart A E, Kwon Y T, Arfin S M, Bradshaw R A, Jenkins N A, Copeland N J, Varshavsky A. J Biol Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 19.Stewart A E, Arfin S M, Bradshaw R A. J Biol Chem. 1995;270:25–28. doi: 10.1074/jbc.270.1.25. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1996. [Google Scholar]
- 21.Bredt D S, Hwang P M, Glatt C E, Lowenstein C, Reed R R, Snyder S H. Nature (London) 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 22.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M I. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlon R A, Rossant J. Development (Cambridge, UK) 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 24.Copeland N G, Jenkins N A. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 25.Dracopoli N C, Haines J L, Korf B R, Moir T D, Morton C C, Seidman C E, Seidman J G, Smith D R. Current Protocols in Human Genetics. New York: Wiley Interscience; 1994. [Google Scholar]
- 26.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 27.Hannoufa A, Negruk V, Eisner G, Lemieux B. Plant J. 1996;10:459–467. doi: 10.1046/j.1365-313x.1996.10030459.x. [DOI] [PubMed] [Google Scholar]
- 28.Borden K L, Freemont P S. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Peters J M, King R W, Page A M, Hieter P, Kirschner M W. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 30.Huibregtse J M, Scheffner M, Beaudenon S, Howley P. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taban C H, Hondermarck H, Bradshaw R A, Boilly B. Experientia. 1996;52:865–870. doi: 10.1007/BF01938871. [DOI] [PubMed] [Google Scholar]
- 32.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 33.Stewart A. Trends in Genetics Nomenclature Guide. Cambridge, U.K.: Elsevier; 1995. [Google Scholar]