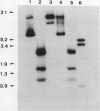
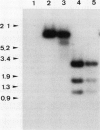
Abstract
The plasmid RSF1010 belongs to a class of plasmids (IncQ) that replicate in a range of bacterial hosts. Although non-self-transmissible, it can be mobilized at high frequency between different gram-negative bacterial species if transfer functions are supplied in trans. We report the transfer of RSF1010 by conjugation from Escherichia coli to the gram-positive actinomycetes Streptomyces lividans and Mycobacterium smegmatis. In its new hosts, the plasmid was stable with respect to structure and inheritance and conferred high-level resistance to streptomycin and sulfonamide. This is the first reported case of conjugative transfer of a naturally occurring plasmid between gram-negative and gram-positive bacteria.
Full text
PDF

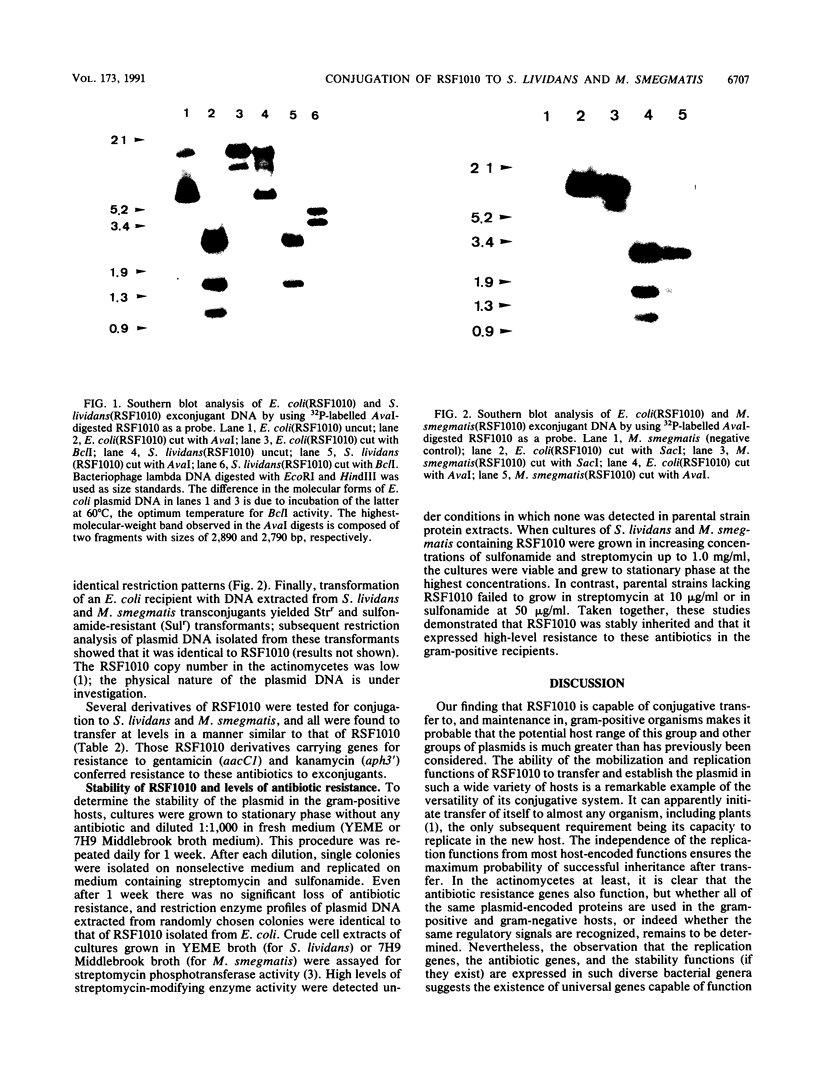

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davison J., Heusterspreute M., Chevalier N., Ha-Thi V., Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51(2-3):275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- Haas M. J., Dowding J. E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Hermans J., Martin C., Huijberts G. N., Goosen T., de Bont J. A. Transformation of Mycobacterium aurum and Mycobacterium smegmatis with the broad host-range gram-negative cosmid vector pJRD215. Mol Microbiol. 1991 Jun;5(6):1561–1566. doi: 10.1111/j.1365-2958.1991.tb00803.x. [DOI] [PubMed] [Google Scholar]
- Jaurin B., Cohen S. N. Streptomyces lividans RNA polymerase recognizes and uses Escherichia coli transcriptional signals. Gene. 1984 Apr;28(1):83–91. doi: 10.1016/0378-1119(84)90090-8. [DOI] [PubMed] [Google Scholar]
- Labes M., Pühler A., Simon R. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene. 1990 Apr 30;89(1):37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- Lazraq R., Clavel-Sérès S., David H. L., Roulland-Dussoix D. Conjugative transfer of a shuttle plasmid from Escherichia coli to Mycobacterium smegmatis [corrected]. FEMS Microbiol Lett. 1990 May;57(1-2):135–138. doi: 10.1016/0378-1097(90)90427-r. [DOI] [PubMed] [Google Scholar]
- Martin C., Timm J., Rauzier J., Gomez-Lus R., Davies J., Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990 Jun 21;345(6277):739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- Mazodier P., Petter R., Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989 Jun;171(6):3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranes M. G., Rauzier J., Lagranderie M., Gheorghiu M., Gicquel B. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a "mini" mycobacterium-Escherichia coli shuttle vector. J Bacteriol. 1990 May;172(5):2793–2797. doi: 10.1128/jb.172.5.2793-2797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Bagdasarian M. M., Scholz P., Lurz R., Rückert B., Bagdasarian M. Replication of the broad host range plasmid RSF1010: requirement for three plasmid-encoded proteins. Proc Natl Acad Sci U S A. 1984 Feb;81(3):654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Kalinowski J., Simon R., Seep-Feldhaus A. H., Pühler A. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol. 1990 Mar;172(3):1663–1666. doi: 10.1128/jb.172.3.1663-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990 Nov;4(11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Torsvik V., Goksøyr J., Daae F. L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]