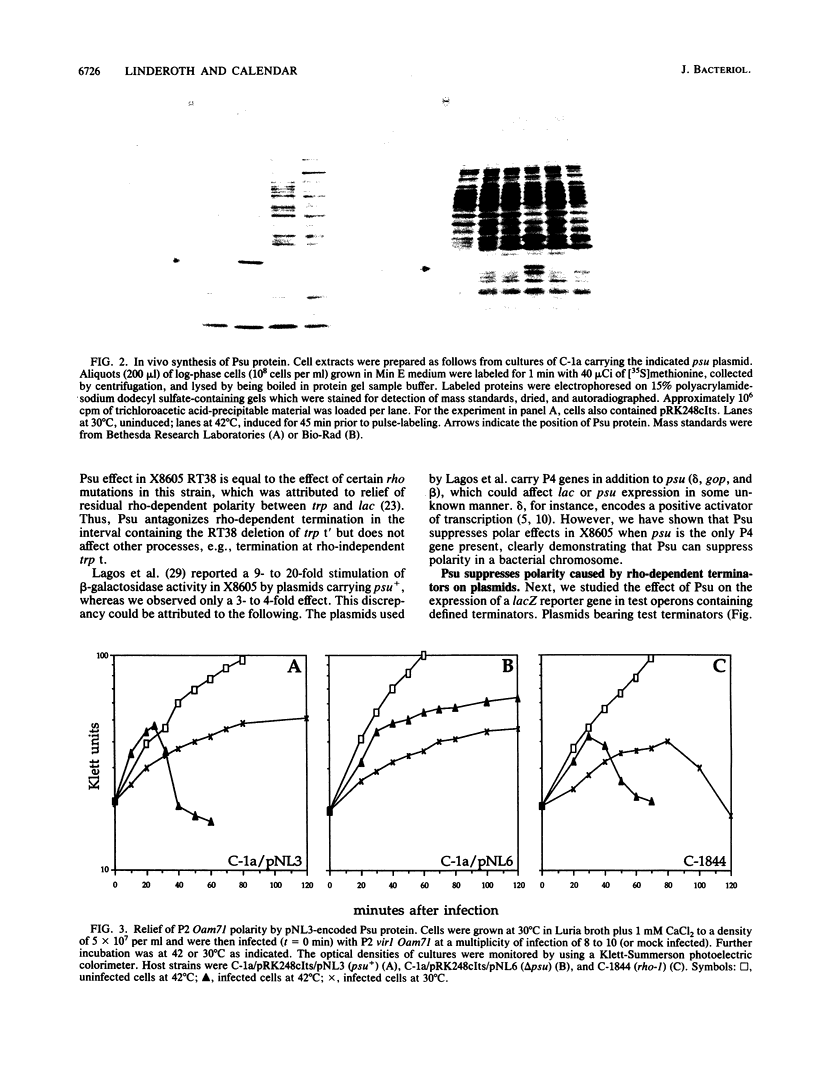
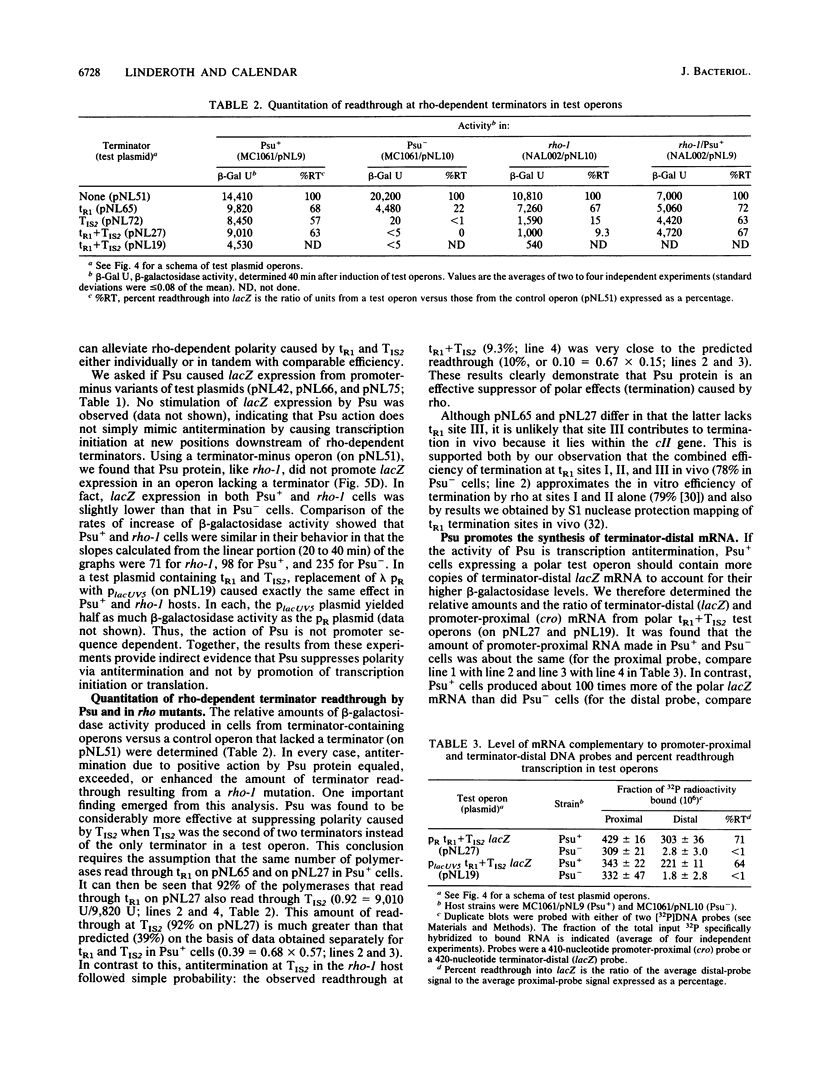
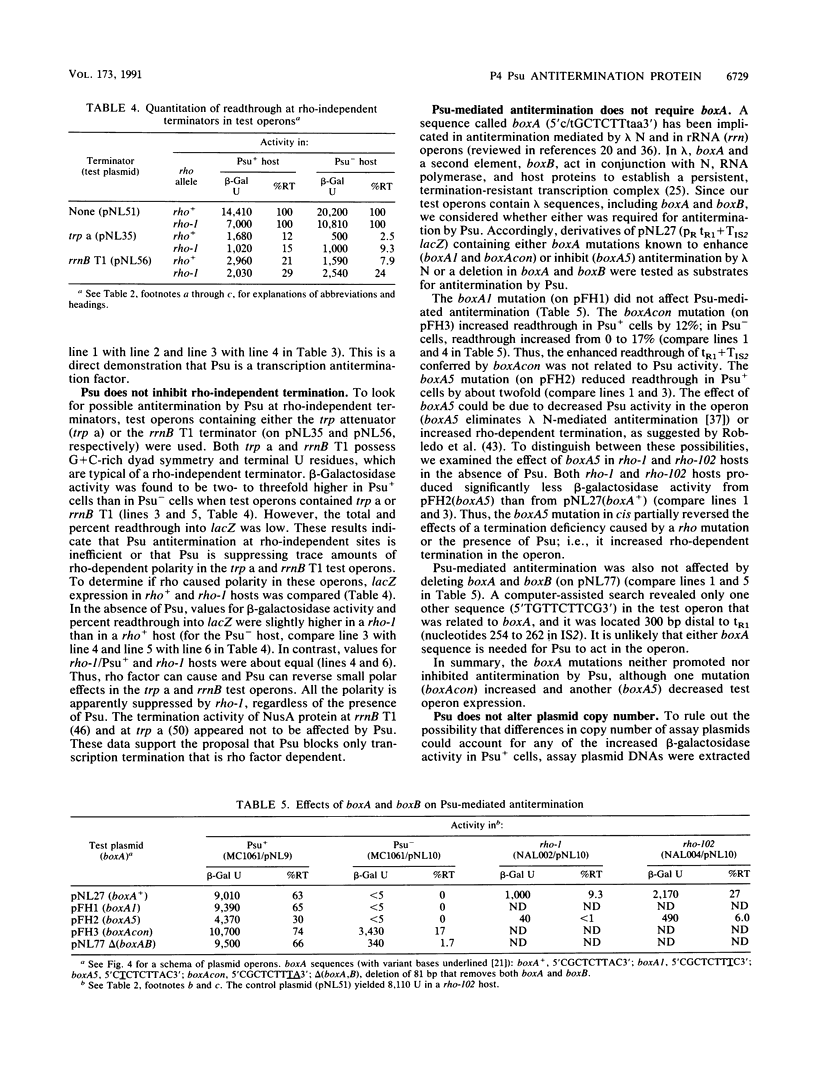
Abstract
A 0.7-kbp DNA fragment from bacteriophage P4 that contained the polarity suppression (psu) gene was cloned in an expression plasmid. Induction of the plasmid-borne psu gene resulted in the overproduction of a protein having the biological properties of the P4-induced polarity suppressor. In vivo, Psu protein acted in trans to suppress rho-dependent polarity in the late genes of an infecting P2 phage, in plasmid operons, and in the host chromosome. Psu action did not require the presence of other P2 or P4 phage genes. Psu caused efficient readthrough (antitermination) by Escherichia coli RNA polymerase at the rho-dependent terminators tR1 and TIS2, individually and in tandem, but did not affect termination at rho-independent sites. Neither the conserved antitermination sequence boxA nor any unique promoter or utilization sequence was required for Psu activity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Barrett K. J., Marsh M. L., Calendar R. Interactions between a satellite bacteriophage and its helper. J Mol Biol. 1976 Sep 25;106(3):683–707. doi: 10.1016/0022-2836(76)90259-x. [DOI] [PubMed] [Google Scholar]
- Baumberg S., Lovett M. G. Reduced recovery of plasmid transconjugants in crosses with Escherichia coli rho mutant recipients. Plasmid. 1977 Nov;1(1):118–122. doi: 10.1016/0147-619x(77)90014-2. [DOI] [PubMed] [Google Scholar]
- Bernard H. U., Helinski D. R. Use of the lambda phage promoter PL to promote gene expression in hybrid plasmid cloning vehicles. Methods Enzymol. 1979;68:482–492. doi: 10.1016/0076-6879(79)68037-0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G. E., Calendar R. Interactions between satellite bacteriophage P4 and its helpers. Annu Rev Genet. 1990;24:465–490. doi: 10.1146/annurev.ge.24.120190.002341. [DOI] [PubMed] [Google Scholar]
- Crowl R., Seamans C., Lomedico P., McAndrew S. Versatile expression vectors for high-level synthesis of cloned gene products in Escherichia coli. Gene. 1985;38(1-3):31–38. doi: 10.1016/0378-1119(85)90200-8. [DOI] [PubMed] [Google Scholar]
- Dale E. C., Christie G. E., Calendar R. Organization and expression of the satellite bacteriophage P4 late gene cluster. J Mol Biol. 1986 Dec 20;192(4):793–803. doi: 10.1016/0022-2836(86)90029-x. [DOI] [PubMed] [Google Scholar]
- Dambly C., Court D., Brachet P. Specificity of polarity suppression in E. coli: correction of defects in gene N, but not in gene Q, of phage lambda. Mol Gen Genet. 1976 Oct 18;148(2):175–182. doi: 10.1007/BF00268383. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Merril C., Adhya S. Interaction of RNA polymerase and rho in transcription termination: coupled ATPase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4828–4832. doi: 10.1073/pnas.75.10.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehó G., Zangrossi S., Ghisotti D., Sironi G. Alternative promoters in the development of bacteriophage plasmid P4. J Virol. 1988 May;62(5):1697–1704. doi: 10.1128/jvi.62.5.1697-1704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. W., Vaughn V., Walter W. A., Neidhardt F. C., Gross C. A. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1987 Jul;1(5):419–432. doi: 10.1101/gad.1.5.419. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Johnson L. L., Alessi D., Craven M. G. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev. 1990 Dec;4(12A):2210–2222. doi: 10.1101/gad.4.12a.2210. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Sommer H., Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979 Mar;6(3):1111–1122. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. P., Mitchell D. H., Beckwith J. Transcription termination at the end of the tryptophan operon of Escherichia coli. J Mol Biol. 1977 May 25;112(3):423–436. doi: 10.1016/s0022-2836(77)80190-3. [DOI] [PubMed] [Google Scholar]
- Halling C., Calendar R., Christie G. E., Dale E. C., Dehò G., Finkel S., Flensburg J., Ghisotti D., Kahn M. L., Lane K. B. DNA sequence of satellite bacteriophage P4. Nucleic Acids Res. 1990 Mar 25;18(6):1649–1649. doi: 10.1093/nar/18.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Kahn M. L., Ziermann R., Dehò G., Ow D. W., Sunshine M. G., Calendar R. Bacteriophage P2 and P4. Methods Enzymol. 1991;204:264–280. doi: 10.1016/0076-6879(91)04013-e. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Yanofsky C. Polarity suppressors increase expression of the wild-type tryptophan operon of Escherichia coli. J Mol Biol. 1976 May 15;103(2):395–409. doi: 10.1016/0022-2836(76)90319-3. [DOI] [PubMed] [Google Scholar]
- Lagos R., Jiang R. Z., Kim S., Goldstein R. Rho-dependent transcription termination of a bacterial operon is antagonized by an extrachromosomal gene product. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9561–9565. doi: 10.1073/pnas.83.24.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W., Wu R. Transcription terminates at lambda tR1 in three clusters. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6171–6175. doi: 10.1073/pnas.79.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Six E. W. Replication of bacteriophage P4 DNA in a nonlysogenic host. Virology. 1971 Jan;43(1):1–7. doi: 10.1016/0042-6822(71)90218-2. [DOI] [PubMed] [Google Scholar]
- Morgan E. A. Antitermination mechanisms in rRNA operons of Escherichia coli. J Bacteriol. 1986 Oct;168(1):1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. R., Tomich C. S., Friedman D. I. The nusA recognition site. Alteration in its sequence or position relative to upstream translation interferes with the action of the N antitermination function of phage lambda. J Mol Biol. 1984 Dec 25;180(4):1053–1063. doi: 10.1016/0022-2836(84)90270-5. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P., Carey J. L., 3rd rho Factors from polarity suppressor mutants with defects in their RNA interactions. J Biol Chem. 1982 May 25;257(10):5767–5771. [PubMed] [Google Scholar]
- Richardson J. P. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991 Mar 22;64(6):1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Robledo R., Gottesman M. E., Weisberg R. A. Lambda nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990 Apr 20;212(4):635–643. doi: 10.1016/0022-2836(90)90226-c. [DOI] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Sauer B., Ow D., Ling L., Calendar R. Mutants of satellite bacteriophage P4 that are defective in the suppression of transcriptional polarity. J Mol Biol. 1981 Jan 5;145(1):29–46. doi: 10.1016/0022-2836(81)90333-8. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987 Jun 20;195(4):809–818. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- Sunshine M. G., Thorn M., Gibbs W., Calendar R., Kelly B. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology. 1971 Dec;46(3):691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- Sunshine M., Six E. Relief of P2 bacteriophage amber mutant polarity by the satellite bacteriophage P4. J Mol Biol. 1976 Sep 25;106(3):673–682. doi: 10.1016/0022-2836(76)90258-8. [DOI] [PubMed] [Google Scholar]
- Tessman I., Fassler J. S., Bennett D. C. Relative map location of the rep and rho genes of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1637–1640. doi: 10.1128/jb.151.3.1637-1640.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. F., Gottesman M. E. The nus mutations affect transcription termination in Escherichia coli. Nature. 1981 Jul 16;292(5820):212–215. doi: 10.1038/292212a0. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Platt T., Crawford I. P., Nichols B. P., Christie G. E., Horowitz H., VanCleemput M., Wu A. M. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981 Dec 21;9(24):6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Elseviers D., Stauffer G. V., Yanofsky C. Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5988–5992. doi: 10.1073/pnas.75.12.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]



