Abstract
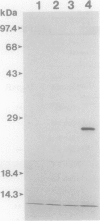
The genes for the peripheral glycerol carbon metabolic pathway (glp) in Pseudomonas aeruginosa are postulated to be positively regulated by GlpR. A gene complementing the glpR2 allele, affecting expression of the putative activator, was cloned by a bacteriophage mini-D3112-based in vivo cloning method. Mini-D3112 replicons were isolated by transfecting glpR2 strain PRP406 and selecting clones able to grow on minimal medium containing glycerol as the sole carbon and energy source. Preliminary biochemical characterization indicated that the cloned activator gene for glycerol metabolism (agmR) may not be allelic to glpR. Restriction analysis and recloning of DNA fragments located the agmR gene to a 2.3-kb EcoRV-SstI DNA fragment. In a T7 RNA polymerase expression system, a single 26,000-Da protein was expressed from this DNA fragment. The amino acid sequence of this protein, deduced from the nucleotide sequence reported here, demonstrates its homology to the effector (or regulator) proteins of the environmentally responsive two-component regulators. The carboxy-terminal region of AgmR contains a possible helix-turn-helix DNA-binding motif and resembles sequences found in transcriptional regulators of the LuxR family.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anba J., Bidaud M., Vasil M. L., Lazdunski A. Nucleotide sequence of the Pseudomonas aeruginosa phoB gene, the regulatory gene for the phosphate regulon. J Bacteriol. 1990 Aug;172(8):4685–4689. doi: 10.1128/jb.172.8.4685-4689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P. C., Vanags R. I., Chakrabarty A. M., Maitra P. K. Fructose 1,6-bisphosphate aldolase activity is essential for synthesis of alginate from glucose by Pseudomonas aeruginosa. J Bacteriol. 1985 Jan;161(1):458–460. doi: 10.1128/jb.161.1.458-460.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Cangelosi G. A., Ankenbauer R. G., Nester E. W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau D. E., Ikenaka K., Tsung K. L., Inouye M. Primary characterization of the protein products of the Escherichia coli ompB locus: structure and regulation of synthesis of the OmpR and EnvZ proteins. J Bacteriol. 1985 Nov;164(2):578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskey S. M., Phibbs P. V., Jr Chromosomal mapping of mutations affecting glycerol and glucose catabolism in Pseudomonas aeruginosa PAO. J Bacteriol. 1985 Jun;162(3):872–880. doi: 10.1128/jb.162.3.872-880.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Casadaban M. J. In vivo cloning of Pseudomonas aeruginosa genes with mini-D3112 transposable bacteriophage. J Bacteriol. 1989 Jul;171(7):3917–3925. doi: 10.1128/jb.171.7.3917-3925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Dikshit R., Konyecsni W. M., Chakrabarty A. M., Misra T. K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989 Mar;171(3):1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRita V. J., Mekalanos J. J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991 Jan 11;64(1):29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M. J., Kadner R. J. Nucleotide sequence of the uhp region of Escherichia coli. J Bacteriol. 1987 Aug;169(8):3556–3563. doi: 10.1128/jb.169.8.3556-3563.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Wallace J. C., Brown J. P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1990;183:111–132. doi: 10.1016/0076-6879(90)83009-x. [DOI] [PubMed] [Google Scholar]
- Igo M. M., Ninfa A. J., Stock J. B., Silhavy T. J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989 Nov;3(11):1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Matsuda Z., Fujiwara T., Lin E. C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990 May;4(5):715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- Kornacki J. A., Burlage R. S., Figurski D. H. The kil-kor regulon of broad-host-range plasmid RK2: nucleotide sequence, polypeptide product, and expression of regulatory gene korC. J Bacteriol. 1990 Jun;172(6):3040–3050. doi: 10.1128/jb.172.6.3040-3050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson T. J., Ye S. Z., Weissenborn D. L., Hoffmann H. J., Schweizer H. Purification and characterization of the repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K12. J Biol Chem. 1987 Nov 25;262(33):15869–15874. [PubMed] [Google Scholar]
- Lessie T. G., Phibbs P. V., Jr Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–388. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- Lessie T. G., Wyk J. C. Multiple forms of Pseudomonas multivorans glucose-6-phosphate and 6-phosphogluconate dehydrogenases: differences in size, pyridine nucleotide specificity, and susceptibility to inhibition by adenosine 5'-triphosphate. J Bacteriol. 1972 Jun;110(3):1107–1117. doi: 10.1128/jb.110.3.1107-1117.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Makino K., Shinagawa H., Amemura M., Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J Mol Biol. 1986 Jul 5;190(1):37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miller J. F., Mekalanos J. J., Falkow S. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science. 1989 Feb 17;243(4893):916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Russell N. J., Gacesa P. Chemistry and biology of the alginate of mucoid strains of Pseudomonas aeruginosa in cystic fibrosis. Mol Aspects Med. 1988;10(1):1–91. doi: 10.1016/0098-2997(88)90002-7. [DOI] [PubMed] [Google Scholar]
- Schweizer H. P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991 Jan 2;97(1):109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- Schweizer H. P. Improved broad-host-range lac-based plasmid vectors for the isolation and characterization of protein fusions in Pseudomonas aeruginosa. Gene. 1991 Jul 15;103(1):87–92. doi: 10.1016/0378-1119(91)90396-s. [DOI] [PubMed] [Google Scholar]
- Schweizer H. P. The pUC18CM plasmids: a chloramphenicol resistance gene cassette for site-directed insertion and deletion mutagenesis in Escherichia coli. Biotechniques. 1990 Jun;8(6):612-3, 616. [PubMed] [Google Scholar]
- Seki T., Yoshikawa H., Takahashi H., Saito H. Cloning and nucleotide sequence of phoP, the regulatory gene for alkaline phosphatase and phosphodiesterase in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):2913–2916. doi: 10.1128/jb.169.7.2913-2916.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Y., Seed B. Electrolyte gradient gels for DNA sequencing. Biotechniques. 1988 Nov-Dec;6(10):942–944. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990 Feb;172(2):659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991 Mar;173(5):1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tsay S. S., Brown K. K., Gaudy E. T. Transport of glycerol by Pseudomonas aeruginosa. J Bacteriol. 1971 Oct;108(1):82–88. doi: 10.1128/jb.108.1.82-88.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991 Feb;173(4):1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]