Abstract
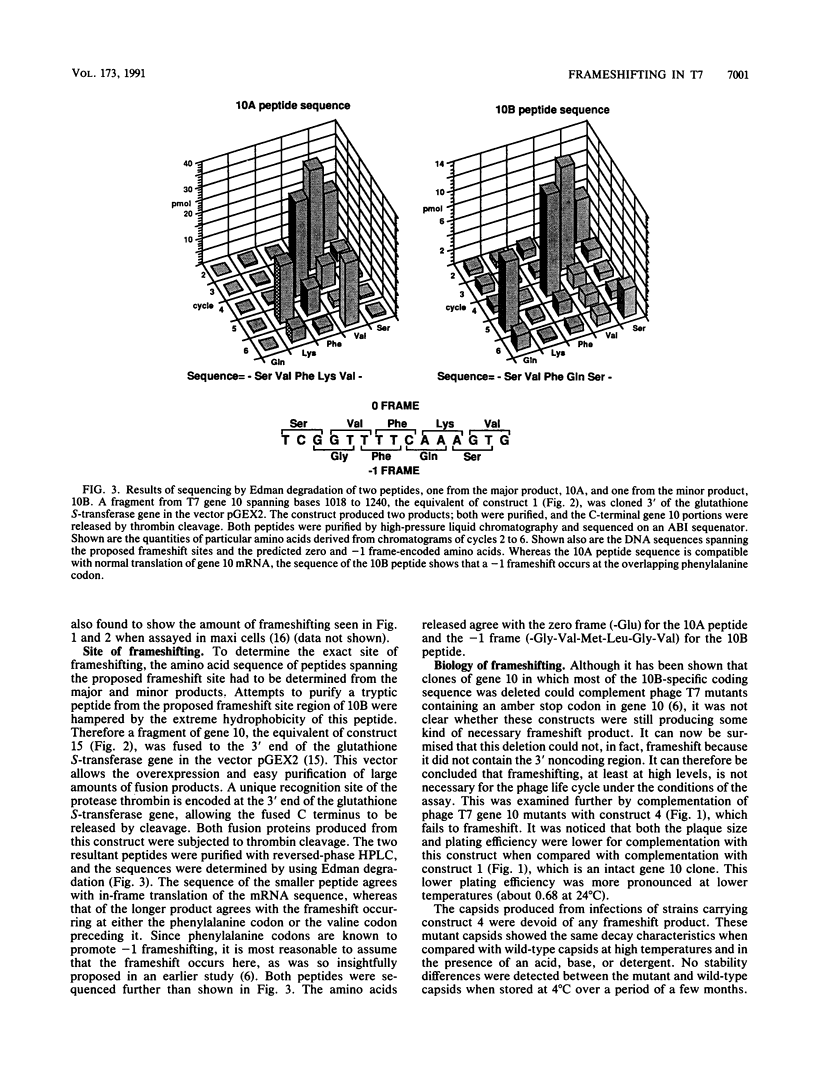
Gene 10 of bacteriophage T7, which encodes the most abundant capsid protein, has two products: a major product, 10A (36 kDa), and a minor product, 10B (41 kDa). 10B is produced by frameshifting into the -1 frame near the end of the 10A coding frame and is incorporated into the capsid. The frameshift occurs at a frequency of about 10% and is conserved in bacteriophage T3. This study shows that sequences important to frameshifting include the originally proposed frameshift site, consisting of overlapping phenylalanine codons and the 3' noncoding region that includes the transcriptional terminator over 200 bases downstream of the frameshift site. The frameshift occurs at the overlapping phenylalanine codons as determined from peptide sequencing data. Complementation studies show that there is only a very weak phenotype associated with phage infections in which there is no 10A frameshifting. Capsids from such infections are devoid of 10B and are as stable as wild-type capsids.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condreay J. P., Wright S. E., Molineux I. J. Nucleotide sequence and complementation studies of the gene 10 region of bacteriophage T3. J Mol Biol. 1989 Jun 5;207(3):555–561. doi: 10.1016/0022-2836(89)90464-6. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Lee C. C., Caskey C. T. Recent advances in peptide chain termination. Mol Microbiol. 1990 Jun;4(6):861–865. doi: 10.1111/j.1365-2958.1990.tb00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne V., Rao V. B., Black L. W. A bacteriophage T4 DNA packaging related DNA-dependent ATPase-endonuclease. J Biol Chem. 1982 Nov 25;257(22):13223–13232. [PubMed] [Google Scholar]
- Masamune Y., Frenkel G. D., Richardson C. C. A mutant of bacteriophage T7 deficient in polynucleotide ligase. J Biol Chem. 1971 Nov 25;246(22):6874–6879. [PubMed] [Google Scholar]
- Molineux I. J., Schmitt C. K., Condreay J. P. Mutants of bacteriophage T7 that escape F restriction. J Mol Biol. 1989 Jun 5;207(3):563–574. doi: 10.1016/0022-2836(89)90465-8. [DOI] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989 Sep;53(3):273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce J. C., Masker W. E. A single-base change in gene 10 of bacteriophage T7 permits growth on Shigella sonnei. J Virol. 1988 Nov;62(11):4369–4371. doi: 10.1128/jvi.62.11.4369-4371.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipley J., Stassi D., Dunn J., Goldman E. Analysis of bacteriophage T7 gene 10A and frameshifted 10B proteins. Gene Expr. 1991 May;1(2):127–136. [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Relationships among different strains of T7 and among T7-related bacteriophages. Virology. 1979 May;95(1):70–84. doi: 10.1016/0042-6822(79)90402-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H. Genetic and physical mapping of the late region of bacteriophage T7 DNA by use of cloned fragments of T7 DNA. J Mol Biol. 1981 Dec 15;153(3):503–525. doi: 10.1016/0022-2836(81)90405-8. [DOI] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]




