Abstract
As reported previously, Saccharomyces cerevisiae cells deficient in all four known genes coding for alcohol dehydrogenases (ADH1 through ADH4) produce considerable amounts of ethanol during aerobic growth on glucose. It has been suggested that ethanol production in such adh0 cells is a corollary of acetaldehyde dismutation in mitochondria. This could be substantiated further by showing that mitochondrial ethanol formation requires functional electron transport, while the proton gradient or oxidative phosphorylation does not interfere with reduction of acetaldehyde in isolated mitochondria. This acetaldehyde-reducing activity is different from classical alcohol dehydrogenases in that it is associated with the inner mitochondrial membrane and also is unable to carry out ethanol oxidation. The putative cofactor is NADH + H+ generated by a soluble, matrix-located aldehyde dehydrogenase upon acetaldehyde oxidation to acetate. This enzyme has been purified from mitochondria of glucose-grown cells. It is clearly different from the known mitochondrial aldehyde dehydrogenase, which is absent in glucose-grown cells. Both acetaldehyde-reducing and acetaldehyde-oxidizing activities are also present in the mitochondrial fraction of fermentation-proficient (ADH+) cells. Mitochondrial acetaldehyde dismutation may have some significance in the removal of surplus acetaldehyde and in the formation of acetate in mitochondria during aerobic glucose fermentation.
Full text
PDF
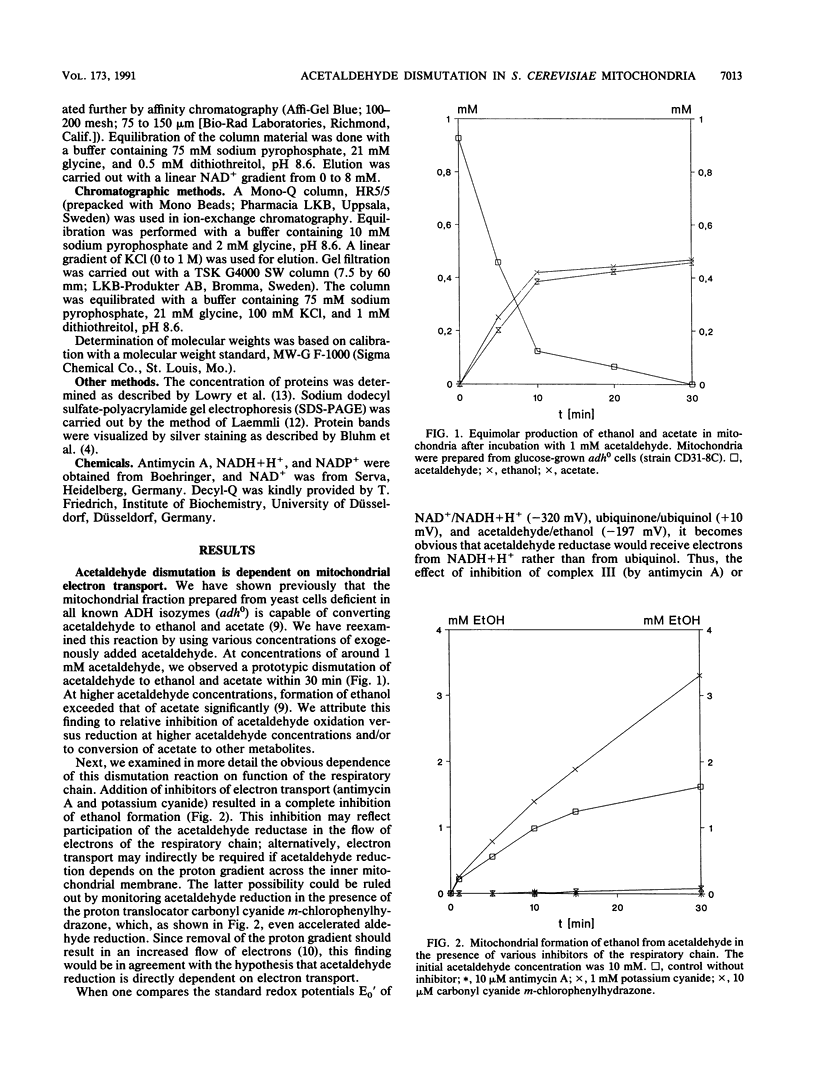
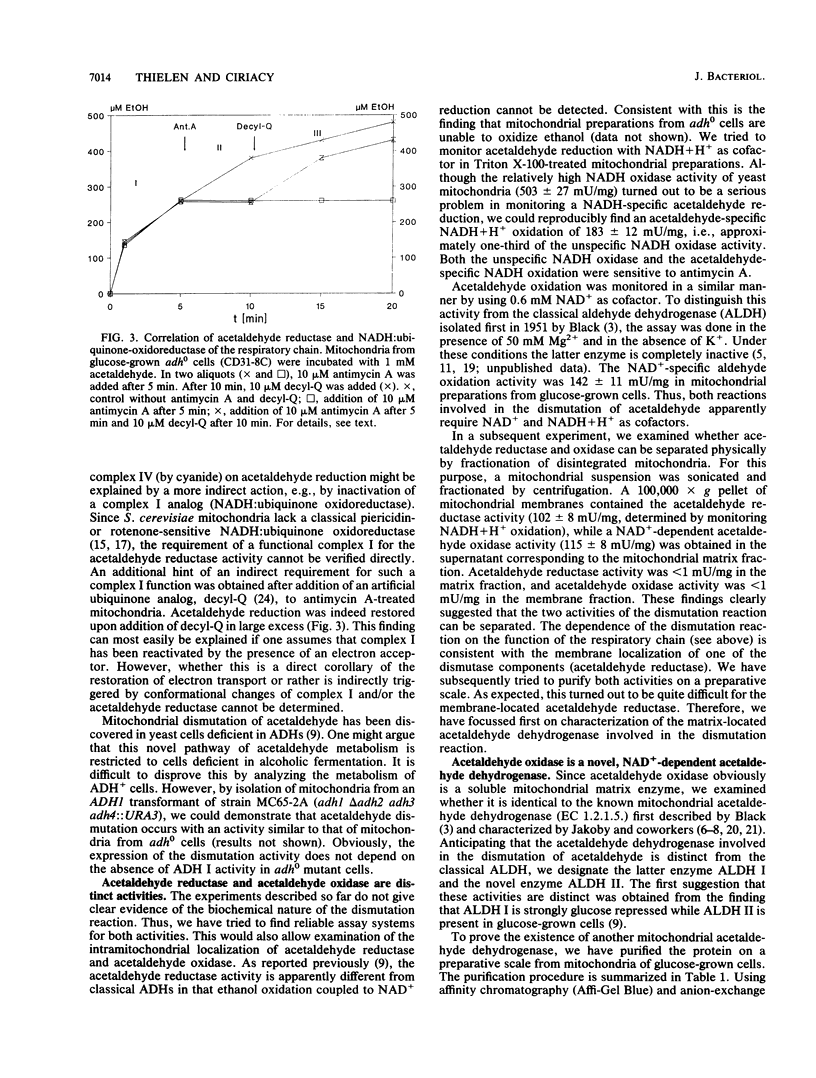
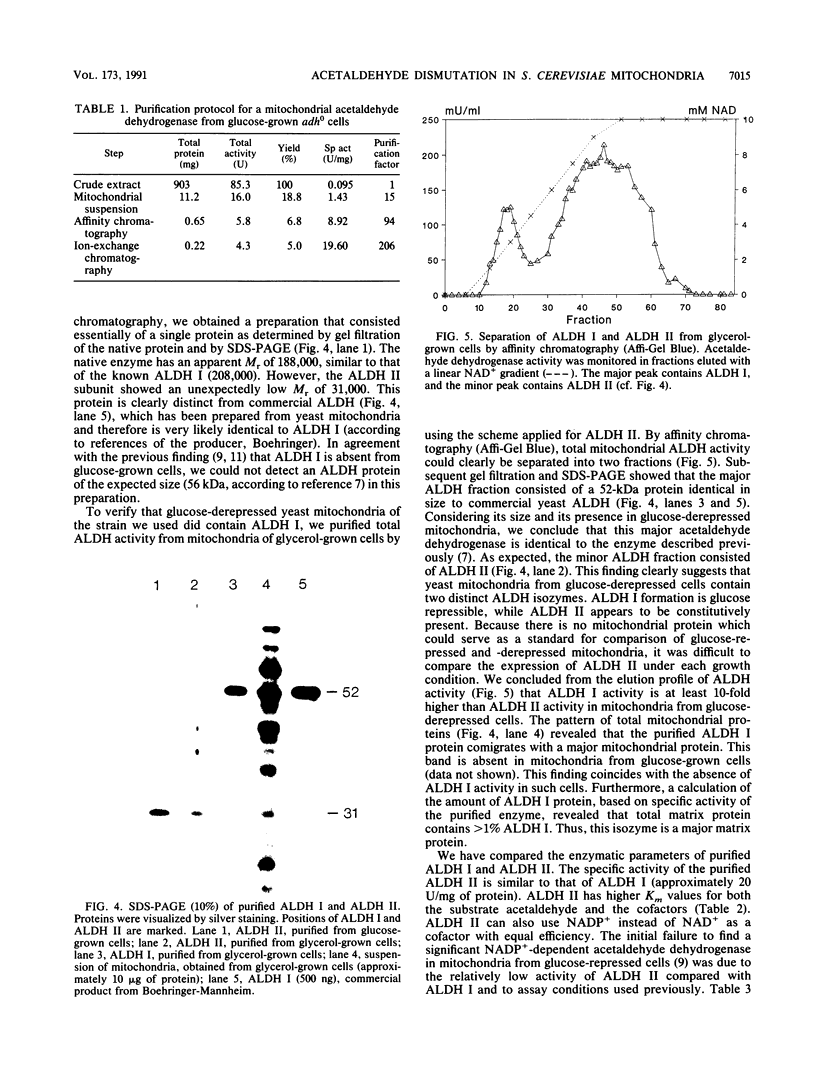


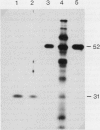
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BLACK S. Yeast aldehyde dehydrogenase. Arch Biochem Biophys. 1951 Nov;34(1):86–97. doi: 10.1016/s0003-9861(51)80013-4. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Betts G. F. Rapid purification and properties of potassium-activated aldehyde dehydrogenase from Saccharomyces cerevisiae. Biochem J. 1978 Sep 1;173(3):773–786. doi: 10.1042/bj1730773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury S. L., Jakoby W. B. Ordered binding of substrates to yeast aldehyde dehydrogenase. J Biol Chem. 1971 Mar 25;246(6):1834–1840. [PubMed] [Google Scholar]
- Clark J. F., Jakoby W. B. Yeast aldehyde dehydrogenase. 3. Preparation of three homogeneous species. J Biol Chem. 1970 Nov 25;245(22):6065–6071. [PubMed] [Google Scholar]
- Clark J. F., Jakoby W. B. Yeast aldehyde dehydrogenase. IV. Dissociation and reassociation of native and hybrid forms. J Biol Chem. 1970 Nov 25;245(22):6072–6077. [PubMed] [Google Scholar]
- Drewke C., Thielen J., Ciriacy M. Ethanol formation in adh0 mutants reveals the existence of a novel acetaldehyde-reducing activity in Saccharomyces cerevisiae. J Bacteriol. 1990 Jul;172(7):3909–3917. doi: 10.1128/jb.172.7.3909-3917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heytler P. G. Uncouplers of oxidative phosphorylation. Methods Enzymol. 1979;55:462–442. doi: 10.1016/0076-6879(79)55060-5. [DOI] [PubMed] [Google Scholar]
- Jacobson M. K., Bernofsky C. Mitochondrial acetaldehyde dehydrogenase from Saccharomyces cerevisiae. Biochim Biophys Acta. 1974 Jun 18;350(2):277–291. doi: 10.1016/0005-2744(74)90502-6. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mackler B., Haynes B. Studies of oxidation phosphorylation in Saccharomyces cerevisiae and Saccharomyces carlsbergensis. Biochim Biophys Acta. 1973 Jan 18;292(1):88–91. doi: 10.1016/0005-2728(73)90253-3. [DOI] [PubMed] [Google Scholar]
- Onishi T. Mechanism of electron transport and energy conservation in the site I region of the respiratory chain. Biochim Biophys Acta. 1973 Dec 7;301(2):105–128. [PubMed] [Google Scholar]
- Pickett M., Gwynne D. I., Buxton F. P., Elliott R., Davies R. W., Lockington R. A., Scazzocchio C., Sealy-Lewis H. M. Cloning and characterization of the aldA gene of Aspergillus nidulans. Gene. 1987;51(2-3):217–226. doi: 10.1016/0378-1119(87)90310-6. [DOI] [PubMed] [Google Scholar]
- Sorger G. J., Evans H. J. Effects of univalent cations on the properties of yeast NAD+ acetaldehyde dehydrogenase. Biochim Biophys Acta. 1966 Apr 12;118(1):1–8. doi: 10.1016/s0926-6593(66)80139-x. [DOI] [PubMed] [Google Scholar]
- Steinman C. R., Jakoby W. B. Yeast aldehyde dehydrogenase. I. Purification and crystallization. J Biol Chem. 1967 Nov 10;242(21):5019–5023. [PubMed] [Google Scholar]
- Steinman C. R., Jakoby W. B. Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations. J Biol Chem. 1968 Feb 25;243(4):730–734. [PubMed] [Google Scholar]
- Wills C., Phelps J. A technique for the isolation of yeast alcohol dehydrogenase mutants with altered substrate specificity. Arch Biochem Biophys. 1975 Apr;167(2):627–637. doi: 10.1016/0003-9861(75)90506-8. [DOI] [PubMed] [Google Scholar]
- Zweck A., Bechmann G., Weiss H. The pathway of the quinol/quinone transhydrogenation reaction in ubiquinol: cytochrome-c reductase of Neurospora mitochondria. Eur J Biochem. 1989 Jul 15;183(1):199–203. doi: 10.1111/j.1432-1033.1989.tb14913.x. [DOI] [PubMed] [Google Scholar]