Abstract
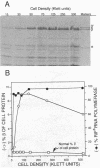
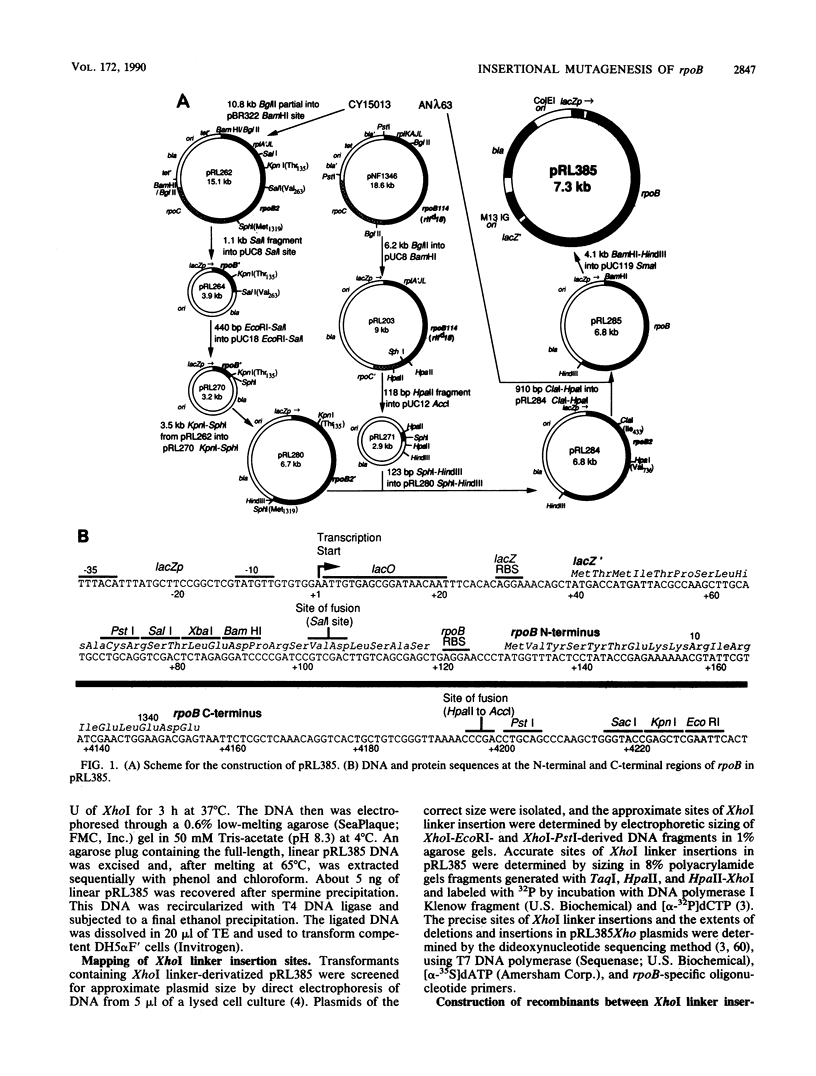
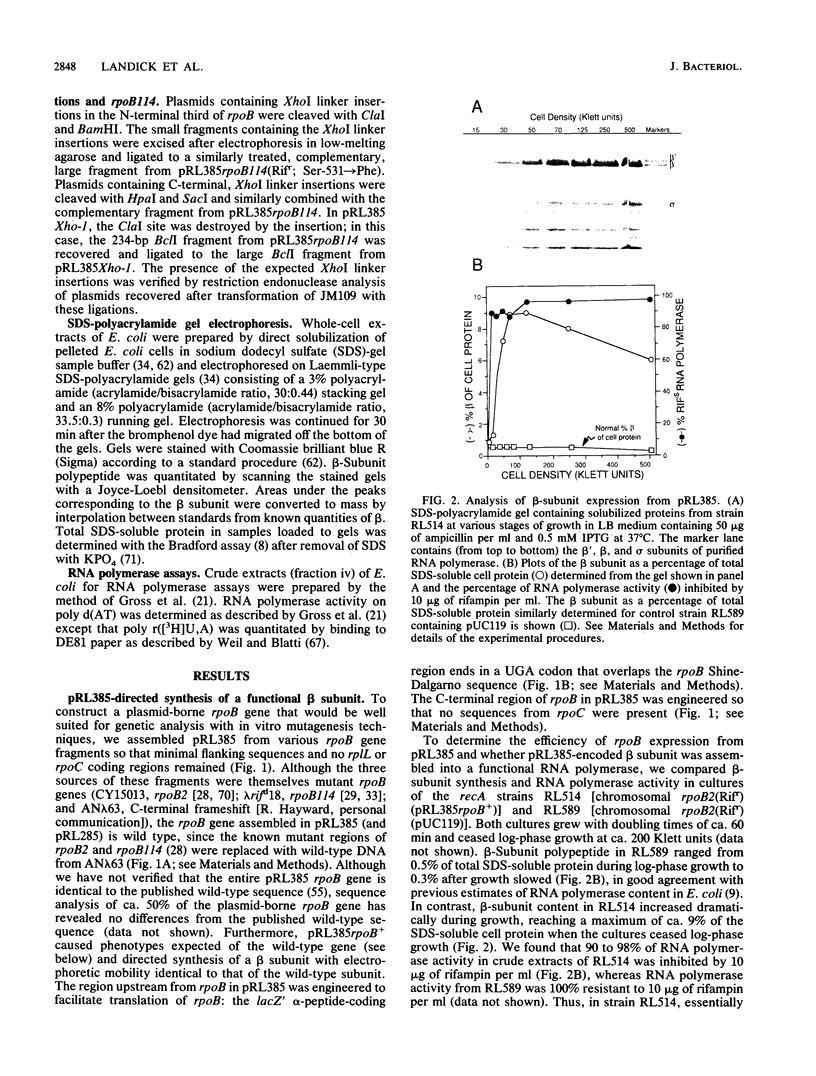
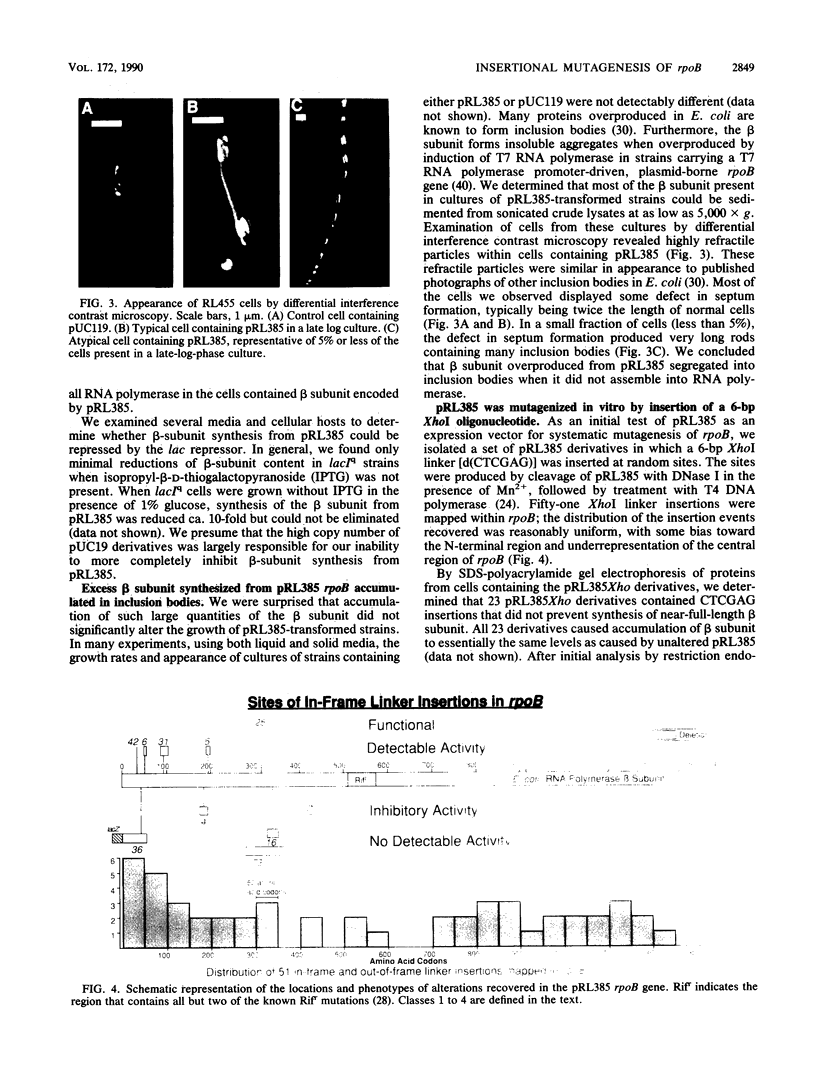
A plasmid was constructed that overproduces the Escherichia coli RNA polymerase beta subunit from a lac promoter-rpoB fusion. The overproduced, plasmid-encoded beta subunit assembled into functional RNA polymerase that supplied greater than 90% of the transcriptional capacity of the cells. Excess beta subunit segregated into insoluble inclusion bodies and was not deleterious to cell growth. By insertion of a XhoI linker sequence (CTCGAG) and accompanying deletion of variable amounts of rpoB sequences, 13 structural alterations were isolated in the first and last thirds of the plasmid-borne rpoB gene. Twelve of these alterations appeared to reduce or prevent assembly of plasmid-encoded beta subunit into RNA polymerase. One alteration had no discernible effect on assembly or function of the beta subunit; eight others appeared to inhibit assembly but still produced detectable transcriptional activity. Three of these nine alterations produced beta-subunit polypeptides that inhibited cell growth at 32 degrees C, even though they were present in less than 50% of the cell RNA polymerase. When assembled into RNA polymerase, these three altered beta subunits apparently affected essential RNA polymerase functions. Four of the recovered alterations appeared to inhibit completely or almost completely assembly of the beta subunit into RNA polymerase. The results are consistent with a hypothesis that sequences in the first third of the beta-subunit polypeptide are especially important for proper folding and assembly of the beta subunit.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S. J., Tittawella I. P., Hayward R. S., Scaife J. G. Amber mutations of Escherichia coli RNA polymerase. Nat New Biol. 1971 Aug 4;232(31):133–136. doi: 10.1038/newbio232133a0. [DOI] [PubMed] [Google Scholar]
- Barnes W. M. Plasmid detection and sizing in single colony lysates. Science. 1977 Jan 28;195(4276):393–394. doi: 10.1126/science.318764. [DOI] [PubMed] [Google Scholar]
- Bedwell D. M., Nomura M. Feedback regulation of RNA polymerase subunit synthesis after the conditional overproduction of RNA polymerase in Escherichia coli. Mol Gen Genet. 1986 Jul;204(1):17–23. doi: 10.1007/BF00330181. [DOI] [PubMed] [Google Scholar]
- Bedwell D., Davis G., Gosink M., Post L., Nomura M., Kestler H., Zengel J. M., Lindahl L. Nucleotide sequence of the alpha ribosomal protein operon of Escherichia coli. Nucleic Acids Res. 1985 Jun 11;13(11):3891–3903. doi: 10.1093/nar/13.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Fiil N. P., Bendiak D., Collins J., Friesen J. D. Expression of Escherichia coli ribosomal protein and RNA polymerase genes cloned on plasmids. Mol Gen Genet. 1979 May 23;173(1):39–50. doi: 10.1007/BF00267689. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Yanofsky C. Mutations of the beta subunit of RNA polymerase alter both transcription pausing and transcription termination in the trp operon leader region in vitro. J Biol Chem. 1983 Jul 10;258(13):8146–8150. [PubMed] [Google Scholar]
- Glass R. E., Honda A., Ishihama A. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. IX. The role of the carboxy-terminus in enzyme assembly. Mol Gen Genet. 1986 Jun;203(3):492–495. doi: 10.1007/BF00422075. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Jones S. T., Ishihama A. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VII. RNA polymerase is a target for ppGpp. Mol Gen Genet. 1986 May;203(2):265–268. doi: 10.1007/BF00333964. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Jones S. T., Nene V., Nomura T., Fujita N., Ishihama A. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VIII. Localisation of a region involved in promoter selectivity. Mol Gen Genet. 1986 Jun;203(3):487–491. doi: 10.1007/BF00422074. [DOI] [PubMed] [Google Scholar]
- Glass R. E., Ralphs N. T., Fujita N., Ishihama A. Assembly of amber fragments of the beta subunit of Escherichia coli RNA polymerase. Eur J Biochem. 1988 Sep 15;176(2):403–407. doi: 10.1111/j.1432-1033.1988.tb14296.x. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Kolocheva T. I., Lukhtanov E. A., Mustaev A. A. Studies on the functional topography of Escherichia coli RNA polymerase. Highly selective affinity labelling by analogues of initiating substrates. Eur J Biochem. 1987 Feb 16;163(1):113–121. doi: 10.1111/j.1432-1033.1987.tb10743.x. [DOI] [PubMed] [Google Scholar]
- Gross C., Engbaek F., Flammang T., Burgess R. Rapid micromethod for the purification of Escherichia coli ribonucleic acid polymerase and the preparation of bacterial extracts active in ribonucleic acid synthesis. J Bacteriol. 1976 Oct;128(1):382–389. doi: 10.1128/jb.128.1.382-389.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C., Hoffman J., Ward C., Hager D., Burdick G., Berger H., Burgess R. Mutation affecting thermostability of sigma subunit of Escherichia coli RNA polymerase lies near the dnaG locus at about 66 min on the E. coli genetic map. Proc Natl Acad Sci U S A. 1978 Jan;75(1):427–431. doi: 10.1073/pnas.75.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M. M., Meares C. F. Topography of transcription: path of the leading end of nascent RNA through the Escherichia coli transcription complex. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4238–4242. doi: 10.1073/pnas.80.14.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Promoter selectivity of prokaryotic RNA polymerases. Trends Genet. 1988 Oct;4(10):282–286. doi: 10.1016/0168-9525(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Transcription signals and factors in Escherichia coli. Adv Biophys. 1986;21:163–173. doi: 10.1016/0065-227x(86)90021-3. [DOI] [PubMed] [Google Scholar]
- Jaskunas S. R., Burgess R. R., Nomura M. Identification of a gene for the alpha-subunit of RNA polymerase at the str-spc region of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5036–5040. doi: 10.1073/pnas.72.12.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988 Jul 5;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Walter W. A., Gross C. A. Characterization of the termination phenotypes of rifampicin-resistant mutants. J Mol Biol. 1988 Jul 20;202(2):245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- Kashlev M. V., Gragerov A. I., Nikiforov V. G. Vliianie reaktsii teplovogo shoka na fenotipicheskoe proiavlenie mutatsii ustoichivosti k rifampitsinu, zatragivaiushchikh gen beta-sub''edinitsy RNK-polimerazy, postavlennoi pod kontrol' laktoznogo promotora. Genetika. 1988 Aug;24(8):1343–1352. [PubMed] [Google Scholar]
- Kawakami K., Ishihama A. Defective assembly of ribonucleic acid polymerase subunits in a temperature-sensitive alpha-subunit mutant of Escherichia coli. Biochemistry. 1980 Jul 22;19(15):3491–3495. doi: 10.1021/bi00556a013. [DOI] [PubMed] [Google Scholar]
- Kirschbaum J. B., Konrad E. B. Isolation of a specialized lambda transducing bacteriophage carrying the beta subunit gene for Escherichia coli ribonucleic acid polymerase. J Bacteriol. 1973 Nov;116(2):517–526. doi: 10.1128/jb.116.2.517-526.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Linn T., Scaife J. Identification of a single promoter in E. coli for rplJ, rplL and rpoBC. Nature. 1978 Nov 2;276(5683):33–37. doi: 10.1038/276033a0. [DOI] [PubMed] [Google Scholar]
- Lisitsyn N. A., Monastyrskaya G. S., Sverdlov E. D. Genes coding for RNA polymerase beta subunit in bacteria. Structure/function analysis. Eur J Biochem. 1988 Nov 1;177(2):363–369. doi: 10.1111/j.1432-1033.1988.tb14385.x. [DOI] [PubMed] [Google Scholar]
- Lisitsyn N. A., Sverdlov E. D., Moiseyeva E. P., Danilevskaya O. N., Nikiforov V. G. Mutation to rifampicin resistance at the beginning of the RNA polymerase beta subunit gene in Escherichia coli. Mol Gen Genet. 1984;196(1):173–174. doi: 10.1007/BF00334112. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney J. D., Lee J. Y., O'Neill R. E., Goldfarb A. Overexpression and purification of a biologically active rifampicin-resistant beta subunit of Escherichia coli RNA polymerase. Gene. 1987;58(1):13–18. doi: 10.1016/0378-1119(87)90024-2. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Osawa T., Yura T. Chromosomal location of a structural gene for the RNA polymerase sigma factor in Escherichia coli. Proc Natl Acad Sci U S A. 1977 May;74(5):1831–1835. doi: 10.1073/pnas.74.5.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff N. F., Chamberlin M. J. Termination of transcription by Escherichia coli ribonucleic acid polymerase in vitro. Effect of altered reaction conditions and mutations in the enzyme protein on termination with T7 and T3 deoxyribonucleic acids. Biochemistry. 1980 Jun 24;19(13):3005–3015. doi: 10.1021/bi00554a027. [DOI] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. I. The effect of known, single amino acid substitutions in an essential protein. Mol Gen Genet. 1982;188(3):399–404. doi: 10.1007/BF00330040. [DOI] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. II. Evidence that large N-terminal amber fragments of the beta subunit interfere with RNA polymerase function. Mol Gen Genet. 1982;188(3):405–409. doi: 10.1007/BF00330041. [DOI] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. IV. Structure-function correlates. Mol Gen Genet. 1984;194(1-2):166–172. doi: 10.1007/BF00383512. [DOI] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. VI. A redundant region in the beta polypeptide. Mol Gen Genet. 1984;196(1):64–67. doi: 10.1007/BF00334093. [DOI] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Relaxed mutants of Escherichia coli RNA polymerase. FEBS Lett. 1983 Mar 21;153(2):307–310. doi: 10.1016/0014-5793(83)80630-9. [DOI] [PubMed] [Google Scholar]
- Newman A., Hayward R. S. Cloning of DNA of the rpoBC operon from the chromosome of Escherichia coli K12. Mol Gen Genet. 1980 Feb;177(3):527–533. doi: 10.1007/BF00271493. [DOI] [PubMed] [Google Scholar]
- Oeschger M. P., Oeschger N. S., Wiprud G. T., Woods S. L. High efficiency temperature-sensitive amber suppressor strains of Escherichia coli K12: isolation of strains with suppressor-enhancing mutations. Mol Gen Genet. 1980;177(4):545–552. doi: 10.1007/BF00272662. [DOI] [PubMed] [Google Scholar]
- Oeschger M. P., Wiprud G. T. High efficiency temperature-sensitive amber suppressor strains of Escherichia coli K12: construction and characterization of recombinant strains with suppressor-enhancing mutations. Mol Gen Genet. 1980;178(2):293–299. doi: 10.1007/BF00270475. [DOI] [PubMed] [Google Scholar]
- Ohme M., Tanaka M., Chunwongse J., Shinozaki K., Sugiura M. A tobacco chloroplast DNA sequence possibly coding for a polypeptide similar to E. coli RNA polymerase beta-subunit. FEBS Lett. 1986 May 5;200(1):87–90. doi: 10.1016/0014-5793(86)80516-6. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Salomatina I. S., Shuvaeva T. M., Lipkin V. M., Sverdlov E. D. The primary structure of E. coli RNA polymerase, Nucleotide sequence of the rpoC gene and amino acid sequence of the beta'-subunit. Nucleic Acids Res. 1982 Jul 10;10(13):4035–4044. doi: 10.1093/nar/10.13.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Chertov OYu, Modyanov N. N., Grinkevich V. A., Makarova I. A., Marchenko T. V., Polovnikova I. N. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the beta-subunit. Eur J Biochem. 1981 Jun 1;116(3):621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Monastyrskaya G. S., Guriev S. O., Kalinina N. F., Sverdlov E. D., Gragerov A. I., Bass I. A., Kiver I. F., Moiseyeva E. P., Igumnov V. N. RNA polymerase rifampicin resistance mutations in Escherichia coli: sequence changes and dominance. Mol Gen Genet. 1983;190(2):344–348. doi: 10.1007/BF00330662. [DOI] [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Ridley S. P., Oeschger M. P. An amber mutation in the gene encoding the beta' subunit of Escherichia coli RNA polymerase. J Bacteriol. 1982 Nov;152(2):736–746. doi: 10.1128/jb.152.2.736-746.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz W., Zillig W. Rifampicin inhibition of RNA synthesis by destabilisation of DNA-RNA polymerase-oligonucleotide-complexes. Nucleic Acids Res. 1981 Dec 21;9(24):6889–6906. doi: 10.1093/nar/9.24.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B. The molecular topography of RNA polymerase-promoter interaction. Cell. 1979 Oct;18(2):277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Blatti S. P. Partial purification and properties of calf thymus deoxyribonucleic acid dependent RNA polymerase III. Biochemistry. 1975 Apr 22;14(8):1636–1642. doi: 10.1021/bi00679a015. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Nomura M. Contranscription of genes for RNA polymerase subunits beta and beta' with genes for ribosomal proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3891–3895. doi: 10.1073/pnas.75.8.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Horn V. Rifampin resistance mutations that alter the efficiency of transcription termination at the tryptophan operon attenuator. J Bacteriol. 1981 Mar;145(3):1334–1341. doi: 10.1128/jb.145.3.1334-1341.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman Z., Verwilghen R. L. Quantitation of proteins solubilized in sodium dodecyl sulfate-mercaptoethanol-Tris electrophoresis buffer. Anal Biochem. 1979 Nov 15;100(1):64–69. doi: 10.1016/0003-2697(79)90110-6. [DOI] [PubMed] [Google Scholar]