Abstract
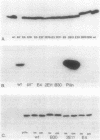
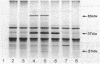
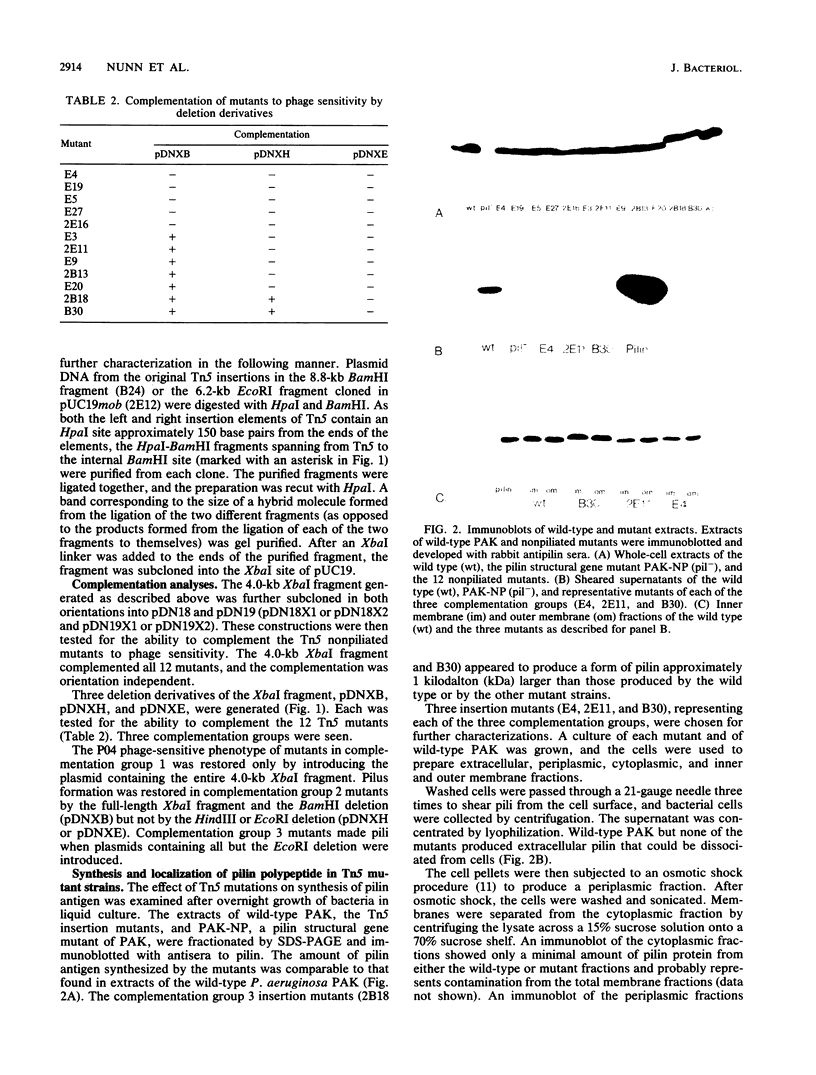
The polar pili of Pseudomonas aeruginosa are composed of monomers of the pilin structural subunits. The biogenesis of pili involves the synthesis of pilin precursor, cleavage of a six-amino-acid leader peptide, membrane translocation, and assembly of monomers into a filamentous structure extending from the bacterial surface. This report describes three novel genes necessary for the formation of pili. DNA sequences adjacent to pilA, the pilin structural gene, were cloned and mutagenized with transposon Tn5. Each of the insertions were introduced into the chromosome of P. aeruginosa PAK by gene replacement. The effect of the Tn5 insertions in the bacterial chromosome on pilus assembly was assessed by electron microscopy and sensitivity of mutants to a pilus-specific bacteriophage. The resultant mutants were also tested for synthesis and membrane localization of the pilin antigen in order to define the genes required for maturation, export, and assembly of pilin. A 4.0-kilobase-pair region of DNA adjacent to the pilin structural gene was found to be essential for formation of pili. This region was sequenced and found to contain three open reading frames coding for 62-, 38- to 45-, and 28- to 32-kilodalton proteins (pilB, pilC, and pilD, respectively). Three proteins of similar molecular weight were expressed in Escherichia coli from the 4.0-kilobase-pair fragment flanking pilA with use of a T7 promoter-polymerase expression system. The results of the analyses of the three genes and the implications for pilin assembly and maturation are discussed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodey G. P., Bolivar R., Fainstein V., Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983 Mar-Apr;5(2):279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. A pilus-dependent Pseudomonas aeruginosa bacteriophage with a long noncontractile tail. Virology. 1973 Feb;51(2):489–492. doi: 10.1016/0042-6822(73)90448-0. [DOI] [PubMed] [Google Scholar]
- Båga M., Normark S., Hardy J., O'Hanley P., Lark D., Olsson O., Schoolnik G., Falkow S. Nucleotide sequence of the papA gene encoding the Pap pilus subunit of human uropathogenic Escherichia coli. J Bacteriol. 1984 Jan;157(1):330–333. doi: 10.1128/jb.157.1.330-333.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig P., Todd T., Sastry P. A., Lee K. K., Hodges R. S., Paranchych W., Irvin R. T. Role of pili in adhesion of Pseudomonas aeruginosa to human respiratory epithelial cells. Infect Immun. 1988 Jun;56(6):1641–1646. doi: 10.1128/iai.56.6.1641-1646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978 Oct;136(1):381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M., Dalrymple B., Delaney S. F., Mattick J. S. Transcription of the fimbrial subunit gene and an associated transfer RNA gene of Pseudomonas aeruginosa. Gene. 1988;62(2):219–227. doi: 10.1016/0378-1119(88)90560-4. [DOI] [PubMed] [Google Scholar]
- Ishimoto K. S., Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Sato T., Yura T. Synthesis and assembly of the membrane proteins in E. coli. Cell. 1977 Jul;11(3):551–559. doi: 10.1016/0092-8674(77)90073-3. [DOI] [PubMed] [Google Scholar]
- Johnson K., Lory S. Characterization of Pseudomonas aeruginosa mutants with altered piliation. J Bacteriol. 1987 Dec;169(12):5663–5667. doi: 10.1128/jb.169.12.5663-5667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Parker M. L., Lory S. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J Biol Chem. 1986 Nov 25;261(33):15703–15708. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leong S. A., Ditta G. S., Helinski D. R. Heme biosynthesis in Rhizobium. Identification of a cloned gene coding for delta-aminolevulinic acid synthetase from Rhizobium meliloti. J Biol Chem. 1982 Aug 10;257(15):8724–8730. [PubMed] [Google Scholar]
- Marrs C. F., Schoolnik G., Koomey J. M., Hardy J., Rothbard J., Falkow S. Cloning and sequencing of a Moraxella bovis pilin gene. J Bacteriol. 1985 Jul;163(1):132–139. doi: 10.1128/jb.163.1.132-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern N. M., O'Donnell I. J., Inglis A. S., Stewart D. J., Clark B. L. Amino acid sequence of pilin from Bacteroides nodosus (strain 198), the causative organism of ovine footrot. FEBS Lett. 1983 Nov 28;164(1):149–153. doi: 10.1016/0014-5793(83)80039-8. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Billyard E., Haas R., Storzbach S., So M. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988 Jun;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Oudega B., De Graaf F. K. Genetic organization and biogenesis of adhesive fimbriae of Escherichia coli. Antonie Van Leeuwenhoek. 1988;54(4):285–299. doi: 10.1007/BF00393521. [DOI] [PubMed] [Google Scholar]
- Pasloske B. L., Paranchych W. The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol Microbiol. 1988 Jul;2(4):489–495. doi: 10.1111/j.1365-2958.1988.tb00055.x. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Saiman L., Ishimoto K., Lory S., Prince A. The effect of piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J Infect Dis. 1990 Mar;161(3):541–548. doi: 10.1093/infdis/161.3.541. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Helinski D. R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S., Lory S. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol. 1986 Feb;165(2):367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S., Lory S. Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J Bacteriol. 1987 Jul;169(7):3181–3188. doi: 10.1128/jb.169.7.3181-3188.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R., Shaw C., Peterson K., Spears P., Mekalanos J. Safe, live Vibrio cholerae vaccines? Vaccine. 1988 Apr;6(2):151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. E., Straus D. C., Johanson W. G., Jr, Berry V. K., Bass J. A. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun. 1980 Sep;29(3):1146–1151. doi: 10.1128/iai.29.3.1146-1151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]