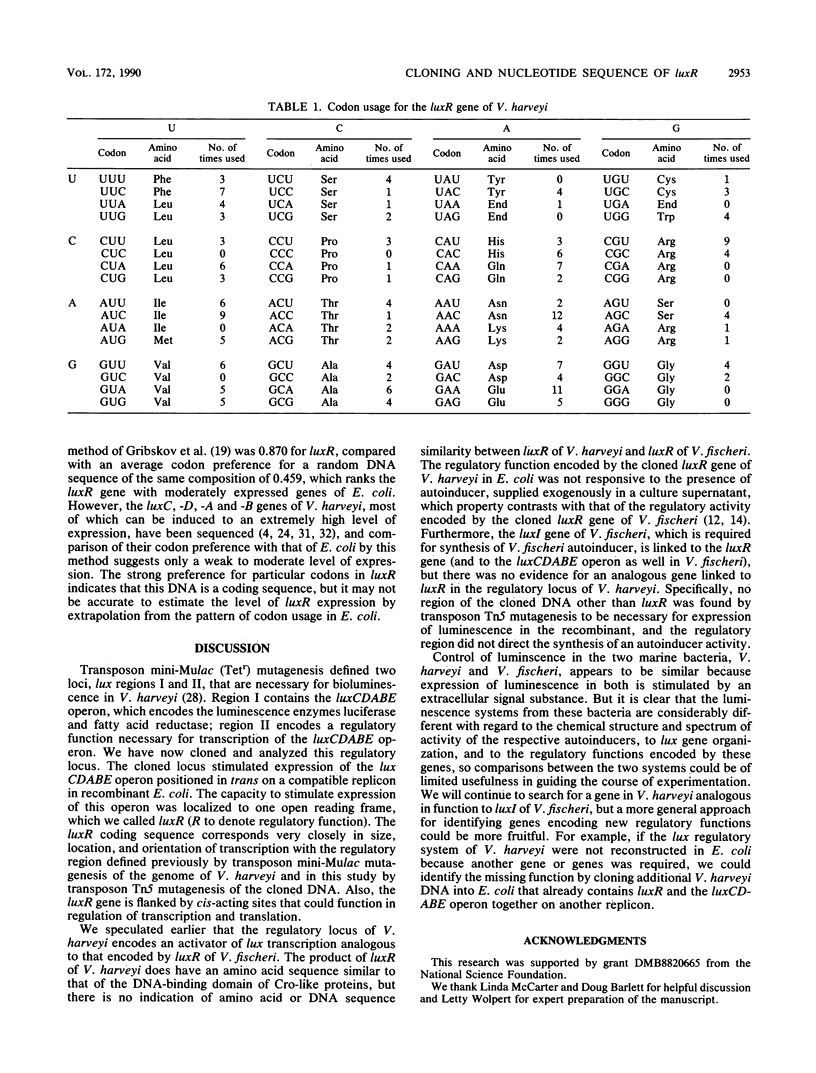
Abstract
Mutagenesis with transposon mini-Mulac was used previously to identify a regulatory locus necessary for expression of bioluminescence genes, lux, in Vibrio harveyi (M. Martin, R. Showalter, and M. Silverman, J. Bacteriol. 171:2406-2414, 1989). Mutants with transposon insertions in this regulatory locus were used to construct a hybridization probe which was used in this study to detect recombinants in a cosmid library containing the homologous DNA. Recombinant cosmids with this DNA stimulated expression of the genes encoding enzymes for luminescence, i.e., the luxCDABE operon, which were positioned in trans on a compatible replicon in Escherichia coli. Transposon mutagenesis and analysis of the DNA sequence of the cloned DNA indicated that regulatory function resided in a single gene of about 0.6-kilobases named luxR. Expression of bioluminescence in V. harveyi and in the fish light-organ symbiont Vibrio fischeri is controlled by density-sensing mechanisms involving the accumulation of small signal molecules called autoinducers, but similarity of the two luminescence systems at the molecular level was not apparent in this study. The amino acid sequence of the LuxR product of V. harveyi, which indicates a structural relationship to some DNA-binding proteins, is not similar to the sequence of the protein that regulates expression of luminescence in V. fischeri. In addition, reconstitution of autoinducer-controlled luminescence in recombinant E. coli, already achieved with lux genes cloned from V. fischeri, was not accomplished with the isolation of luxR from V. harveyi, suggesting a requirement for an additional regulatory component.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belas R., Mileham A., Cohn D., Hilman M., Simon M., Silverman M. Bacterial bioluminescence: isolation and expression of the luciferase genes from Vibrio harveyi. Science. 1982 Nov 19;218(4574):791–793. doi: 10.1126/science.10636771. [DOI] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. G., Meighen E. A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989 Dec 25;264(36):21670–21676. [PubMed] [Google Scholar]
- Cohn D. H., Mileham A. J., Simon M. I., Nealson K. H., Rausch S. K., Bonam D., Baldwin T. O. Nucleotide sequence of the luxA gene of Vibrio harveyi and the complete amino acid sequence of the alpha subunit of bacterial luciferase. J Biol Chem. 1985 May 25;260(10):6139–6146. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Eberhard A., Burlingame A. L., Eberhard C., Kenyon G. L., Nealson K. H., Oppenheimer N. J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981 Apr 28;20(9):2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Eberhard A. Inhibition and activation of bacterial luciferase synthesis. J Bacteriol. 1972 Mar;109(3):1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983 Mar;32(3):773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987 Dec 23;15(24):10455–10467. doi: 10.1093/nar/15.24.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Gotschlich E. C., Seiff M. E., Blake M. S., Koomey M. Porin protein of Neisseria gonorrhoeae: cloning and gene structure. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8135–8139. doi: 10.1073/pnas.84.22.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J. W., Potrikus C. J., Gupta S. C., Kurfürst M., Makemson J. C. Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol. 1985;26:235–291. doi: 10.1016/s0065-2911(08)60398-7. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston T. C., Thompson R. B., Baldwin T. O. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the beta subunit of bacterial luciferase. J Biol Chem. 1986 Apr 15;261(11):4805–4811. [PubMed] [Google Scholar]
- Kempner E. S., Hanson F. E. Aspects of light production by Photobacterium fischeri. J Bacteriol. 1968 Mar;95(3):975–979. doi: 10.1128/jb.95.3.975-979.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Showalter R., Silverman M. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J Bacteriol. 1989 May;171(5):2406–2414. doi: 10.1128/jb.171.5.2406-2414.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Miyamoto C. M., Boylan M., Graham A. F., Meighen E. A. Organization of the lux structural genes of Vibrio harveyi. Expression under the T7 bacteriophage promoter, mRNA analysis, and nucleotide sequence of the luxD gene. J Biol Chem. 1988 Sep 15;263(26):13393–13399. [PubMed] [Google Scholar]
- Miyamoto C. M., Graham A. F., Meighen E. A. Nucleotide sequence of the LuxC gene and the upstream DNA from the bioluminescent system of Vibrio harveyi. Nucleic Acids Res. 1988 Feb 25;16(4):1551–1562. doi: 10.1093/nar/16.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch Microbiol. 1977 Feb 4;112(1):73–79. doi: 10.1007/BF00446657. [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Hastings J. W. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979 Dec;43(4):496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]



