Abstract
Trypanosomatids are parasitic protists that have an ATP-dependent glycolysis with no indication of PPi-dependent metabolism. Most of the glycolysis takes place in peroxisome-like organelles, the glycosomes. We characterized in Trypanosoma brucei a single-copy gene encoding a PPi-dependent enzyme, pyruvate, phosphate dikinase (PPDK), which was expressed functionally in Escherichia coli. Specific antibodies detected a 100-kDa protein in procyclic forms but not in mammalian forms of T. brucei, indicating a differential expression. Glycosomal localization of PPDK was determined by immunofluorescence analysis and was confirmed by Western blot analysis on glycosomal fractions by using anti-PPDK antibodies. Expression and localization of recombinant PPDKs in procyclic forms of T. brucei showed that the AKL motif at the C-terminal extremity of PPDK is necessary for glycosomal targeting. PPDK was detected in every trypanosomatid tested—Trypanosoma congolense, Trypanosoma vivax, Trypanosoma cruzi, Phytomonas, Crithidia and Leishmania—with a good correlation between amount of protein and enzymatic activity. The precise role of PPDK in trypanosomatid carbohydrate metabolism remains to be clarified.
The trypanosomatid family is composed of protists belonging to the order Kinetoplastida, which represents one of the earliest branches in the eukaryotic lineage. They are evolutionarily preceded by amitochondriate protists, such as Giardia lamblia and Trichomonas vaginalis (1). Recent phylogenetic analyses based on rRNA sequences indicate that, among the trypanosomatids, the genus Trypanosoma emerged first, before Phytomonas, followed by a monophyletic clade including Crithidia and Leishmania (2, 3). The genus Trypanosoma is composed of two monophyletic sister groups, the Stercorarian trypanosomes including Trypanosoma cruzi and the Salivarian trypanosomes, also called African trypanosomes. Trypanosoma brucei, Trypanosoma congolense, and Trypanosoma vivax are the most studied representatives of the three subgenera (Trypanozoon, Nannomonas, and Dutonella) that compose the latter group. All of the trypanosomatids are parasites, some of them causing serious diseases in man (Leishmania, T. cruzi, and the African trypanosomes T. brucei gambiense and T. brucei rhodesiense), domestic animals (many African trypanosome species), and plants (Phytomonas).
During their life cycle, African trypanosomes differentiate into several adaptive forms. The most prominent are the bloodstream forms in the mammalian host and the procyclic forms in the midgut of the tsetse fly vector. Adaptation to these different environments requires a concomitant adaptation of their metabolism (4). The procyclic forms use mainly amino acids as an energy source (5) whereas the bloodstream forms rely completely on host-provided glucose to produce ATP (6). In T. congolense and T. vivax, the bloodstream forms produce succinate during glucose metabolism, indicating mitochondrial activity (7) whereas, in T. brucei, the Krebs cycle enzymes are repressed totally and pyruvate is the exclusive end product of glycolysis under aerobic conditions (4).
Glycolysis is a universal metabolic process that shows only minor differences from bacteria to the most complex eukaryotes. One of the most significant deviations from the conventional glycolytic scheme is found in some unicellular protists, such as G. lamblia, T. vaginalis, and Entamoeba histolytica (8, 9). These amitochondriate organisms rely on a phosphofructokinase (PPi-PFK) that uses PPi as phosphoryl donor instead of ATP and contains some or none of the two additional, well characterized PPi-dependent enzymes related to glycolysis [pyruvate, phosphate dikinase (PPDK) and phosphoenolpyruvate (PEP) carboxyphosphotransferase]. The hydrogenosome-bearing type II amitochondriate protists, exemplified by T. vaginalis, have PPi-PFK only whereas the type I organisms (G. lamblia, E. histolytica), without metabolic compartmentation, are the only known protists with PPDK (8, 10). Although PPi-dependent enzymes are able to catalyze reversible reactions, all of the PPi-PFK analyzed so far was characterized in the forward reaction. PPDK in G. lamblia and E. histolytica is assumed to function in the forward reaction (8, 10), though the recent demonstration of pyruvate kinase (PK) in G. lamblia raises some doubt on this point (11).
In contrast, none of the PPi-dependent enzymes have been detected in trypanosomatids in which the first seven glycolytic steps are confined in microbody-like organelles called glycosomes. Glycosomes, like peroxisomes and glyoxysomes, are surrounded by a single lipid-bilayer membrane and are devoid of any DNA. Consequently, all of the glycosomal proteins are nuclear-encoded and contain specific signals responsible for their proper routing to and subsequent uptake by the microbody. Glycolytic enzymes, which are well characterized in trypanosomatids, represent up to 95% of the glycosomal enzyme content, although these organelles also contain enzymes involved in other metabolic pathways present in other microbodies such as fatty acid oxidation and pyrimidine synthesis (6).
Here, we report the characterization of a PPi-dependent enzyme, PPDK, in trypanosomatids. The gene encoding PPDK from T. brucei was cloned and was expressed functionally in Escherichia coli. Subcellular fractionation, Western blotting, and immunofluorescence analyses clearly demonstrate the glycosomal localization of this PPi-dependent enzyme. The role of this enzyme, which was detected in every trypanosomatid we analyzed, remains to be determined.
MATERIALS AND METHODS
Trypanosomatids and Glycosome Preparations.
Long–slender bloodstream forms of T. brucei AnTat1 (Antwerp-Trypanozoon-antigenic-type), 427, and GuTat [kindly provided by A. Seyfang (Oregon Health Sciences University, Portland, OR) and C. E. Davis, University of California, San Diego) were grown in rats while nondividing short–stumpy bloodstream forms of T. brucei GuTat were grown in mice treated with cyclophosphamide as described (12). Bloodstream forms of T. vivax Y481 (isolated at Centre de Recherche sur les Trypanosomiases Animales by Bobo Dioulasso and E. Authié) and T. congolense IL3000 (provided by E. Authié) were grown in irradiated mice. The bloodstream forms were isolated by DEAE ion exchange chromatography as described (13). Procyclic forms of T. brucei 427 and EATRO 1125 [kindly provided by A. Seyfang (Oregon Health Sciences University, Portland, OR) and E. Pays (Université Libre de Brusselles, Brussels) and choanomastigote forms of Crithidia fasciculata (HS6 clone) [kindly provided by E. Tetaud (University of Dundce, Scotland, UK)] were cultured at 27°C in Semi-defined Medium-79 containing 10% (vol/vol) fetal calf serum and 3.5 mg ml−1 hemin (14). Epimastigote and procyclic forms of T. congolense IL3000 (kindly provided by C. Vedrenne and T.B.) were grown at 27°C in Eagle’s minimum essential medium containing 20% (vol/vol) fetal calf serum (15). Epimastigote and metacyclic forms of T. cruzi C.L. [provided by P. Minoprio (Pasteur Institute, Paris)] were cultured and prepared as described (16). Promastigote forms of Leishmania amazonensis were cultured at 26°C in Iscove’s medium supplemented with 10% (vol/vol) fetal calf serum, and amastigote forms were freshly isolated from infected footpads of mice. Phytomonas sp. Euphorbia hyssopifolia (French Guyana) isolate was adapted and grown in Grace’s medium as described elsewhere (2). Glycosomes were prepared from procyclic and bloodstream forms of T. brucei as described (17) after homogenizing the cells with silicon carbide as grinding material (18).
Cloning and Sequencing of the PPDK Gene.
The ptb34 clone was isolated randomly from a T. brucei DiTat1.6 (bloodstream forms) cDNA library. cDNA was synthesized from poly(A)+ mRNA without the usual oligo-dT primer as described (19) and was inserted into the EcoRI site of λZAP II cloning vector (Stratagene). Recombinant pBluescript II plasmid containing the ptb34 fragment was excised from the λZAP II clones as recommended by the manufacturer (Stratagene). The ptb34 fragment was completely sequenced with universal, reverse, and specific primers by the dideoxynucleotide chain termination method.
A T. brucei genomic DNA library, generated into the c2X75 cosmid vector (20) as described (21), was probed with the [32P]-labeled ptb34 cDNA fragment (22). The Cos8 clone was selected and subcloned into pUC18 vector linearized with HindIII and was dephosphorylated (Appligene, Strasbourg, France). Different clones containing HindIII fragments, selected by screening with the [32P]-labeled ptb34 cDNA fragment, were sequenced partially or completely with universal, reverse, and specific primers by the dideoxynucleotide chain termination method by using AmpliTaq DNA polymerase as described by the manufacturer (Perkin–Elmer).
Expression of Recombinant PPDK in Procyclic Forms of T. brucei.
The PPDK gene, containing or not the C-terminal AKL motif and tagged with an N-terminal poly-histidine, was inserted into the pTSA-3′proc expression vector (kindly provided by D. Salmon and E. Pays) designed for gene expression in procyclic forms of T. brucei (Fig. 1). A 5′ oligonucleotide (PPDKpro3) containing a XhoI restriction site (underlined) followed by an ATG codon (bold face), six histidine codons (lower case), and the 5′ end of the PPDK gene (5′-CCAAATCTCGAGACAATGcaccatcaccatcaccatATGGTGGCTAAAAAGTGGGTT-3′) was used together with 2 different 3′ oligonucleotides containing the 3′ end of the PPDK gene with (PPDKpro4) or without (PPDKpro5) the last three codons encoding the AKL motif (lower case) followed by a stop codon (bold face) and a XbaI restriction site (underlined) (PPDKpro4: 5′-CCCACGTCTAGATTAaagcttggcCGCGAATCC-3′; PPDKpro5: 5′-CTTTTTATCTAGATTACGCGAATCCCTTACGC GC-3′) to generate two PCR fragments (PPDK34 and PPDK35) by using the Cos8 cosmid as a matrix and AmpliTaq Gold (Perkin–Elmer) (23). Both XhoI/XbaI-digested PCR fragments were inserted into the pTSA-3′proc vector previously digested by XhoI and XbaI to generate the pTSA-PPDK34 and pTSA-PPDK35 plasmids.
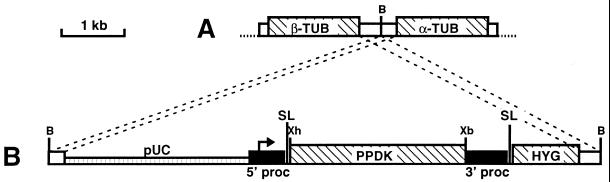
Figure 1.
Map of pTSA-3′proc vector designed for gene expression in procyclic forms of T. brucei (kindly provided by D. Salmon and E. Pays). The coding sequences (PPDK: pyruvate, phosphate dikinase; HYG: hygromycine resistance; α-TUB and β-TUB: α- and β-tubulin) are indicated by hatched boxes. The black boxes flanking the PPDK gene contain the procyclin promoter indicated by an arrow (5′ proc) or the polyadenylation signal present in the 3′ noncoding region of the procyclin (3′ proc), and the splice leaders (SL) are shown. The white boxes represent the tubulin intergenic region used to target the insertion of the recombinant pTSA-3′proc (B) into the tubulin gene cluster (A), and the pUC18 vector is shown as a hatched thin box. Abbreviations: B, BssHII; Xb, XbaI and Xh, XhoI.
107 cells (EATRO1125 procyclic forms) resuspended in 0.5 ml ZPFM (132 mM NaCl/8 mM KCl/8 mM Na2HPO4/1.5 mM KH2PO4/0.5 mM MgAc2/0.09 mM CaAc2) were incubated for 5 min at room temperature with 5 μg of pTSA-PPDK34 or pTSA-PPDK35 BssHII-digested plasmids prepared with Wizard Plus Maxiprep kit (Promega). Then, electroporation was immediately performed in 2-mm electroporation cuvettes with the Cellject Electroporation System apparatus (Eurogentec, Brussels) by using as parameters 1,500 V, 25 μF and an infinite resistance. The cells then were resuspended in 4 ml of Semi-defined Medium-79; 25 μg ml−1 Hygromycin B (Sigma) was added 24 hr later to the culture medium.
Functional Expression and Purification of Recombinant PPDK in E. coli.
Two steps were necessary to insert the PPDK gene containing six histidine residues at the C-terminal extremity between NdeI and BamHI restriction sites of pET3a expression vector (Novagen) because both sites are present in this gene. A NdeI/BamHI- digested PCR fragment containing the 4/5 N-terminal region of the gene was inserted into the pET3a vector previously digested by NdeI and BamHI to generate the pET-PPDK13 recombinant plasmid. Then, a second PCR fragment containing the 2/5 C-terminal region of the gene ending with six histidine codons followed by the stop codon was digested to completion by BstBI and was digested partially by BamHI (another BamHI site, which should not be cleaved, is present within the fragment). The BstBI/BamHI-digested PCR fragment was inserted into the BstBI/BamHI-digested pET-PPDK13 plasmid to yield pET-PPDK15 plasmid that encodes the full-length PPDK containing the poly(His)-tag (PPDK15). Expression and purification of recombinant PPDK15 was performed as described by the manufacturer (Novagen).
PPDK and Glycerol-3-Phosphate Dehydrogenase (GPDH) Activities.
PPDK activity was assayed in both forward (pyruvate production from phosphoenolpyruvate) and reverse (phosphoenolpyruvate production from pyruvate) directions on purified recombinant PPDK from transformed E. coli with the pET-PPDK15 plasmid. Sonicated (5 sec at 4°C) crude extracts of trypanosomatids resuspended in 300 mM NaCl, 50 mM Tris (pH 8), or glycosomal fractions resuspended in buffer (250 ml of sucrose/25 ml Tris (pH 7.4)/1 ml EDTA) were tested for both PPDK and GPDH activities. For the PPDK assay, both pyruvate and PEP formation were measured spectrophotometrically at 340 nm via oxidation of NADH in the presence of a coupling system containing lactate dehydrogenase (24) or PK and lactate dehydrogenase (25), respectively. For the GPDH assay, oxidation of NADH was monitored spectrophotometrically at 340 nm (26).
Production of PPDK Antibodies and Detection of PPDK.
A NdeI/BamHI-digested PCR fragment encoding 180 amino acids of p110 protein (from amino acid 529 to amino acid 856) was inserted into pET3a vector (Novagen) previously digested with NdeI and BamHI. The resulting plasmid, pET-PPDK43, encodes a 20-kDa peptide corresponding to the PPDK fragment (180 aa) plus six histidine residues at the N-terminal extremity preceeded by an ATG codon (PPDK43). Expression and purification of the recombinant PPDK43 peptide was performed as described by the manufacturer (Novagen). Antisera were raised in rabbit and mice by five injections at 2-week intervals of 100 μg and 20 μg, respectively, of PPDK43 peptide (electroeluted after separation by SDS/PAGE) emulsified with complete Freund adjuvant (first injection) or incomplete Freund adjuvant. Furthermore, mAbs were developed as described (27). Two anti-PPDK antibodies producing hybridomas were selected: H112 was specific for T. brucei PPDK whereas H122 recognized the PPDK from every tested trypomanosomatid so far.
For Western blots, total extracts of trypanosomatids or glycosomal fractions from procyclic or bloodstream forms of T. brucei were boiled for 2 min in 1% SDS. Sample preparation, migration in SDS-polyacrylamide gels, immunoblotting on Immobilon-P filters (Millipore) and immunodetection using, as secondary antibody, anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad) were carried out as described (22, 28). Mouse anti-PPDK43 peptide or rabbit anti-GPDH (H. Denise, C. Giroud, and T.B., unpublished material) and mAbs anti-poly(His) (Novagen) were diluted 1:500. Hybridoma culture supernatants containing anti-PPDK (H112 or H122) were used undiluted.
For immunolocalization, log phase cells were fixed with 2% formaldehyde in PBS (0.15 M NaCl/5 mM K-phosphate, pH 7.4) for 30 min at 25°C. The solution was adjusted to 0.1% Triton X-100 and was incubated for 10 min; then, glycine (0.1 M) was added for 10 min to neutralize active aldehyde groups. After centrifugation, the cells were resuspended in PBS and allowed to adhere to glass slides until completely dry. The slides then were incubated for 30 min with different antibodies. The fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody diluted 1:100 in PBS plus 0.1% BSA and 0.1% Triton X-100 was added for 30 min. The slides were washed with PBS and mounted with antifade solution (Vectashield, Vector Laboratories). The cells were observed with a Zeiss UV microscope, and images were captured by camera (Photometrics, Tucson, AZ) and iplab software (Signal Analytics, Vienna) and were merged in Adobe photoshop (Adobe Systems, Mountain View, CA) on a Macintosh 7100/80 computer.
RESULTS
Cloning and Differential Expression of T. brucei PPDK Gene.
A cloned cDNA fragment (ptb34) containing an ORF encoding a peptide with a high degree of identity with PPDKs was isolated from a cDNA library (data not shown). We used the ptb34 fragment as probe to isolate from a cosmid library of T. brucei AnTat1 (bloodstream forms) a genomic fragment (Cos8 clone) that contains the full-length gene. We determined that this single copy gene (data not shown) encodes a protein of 912 amino acids (Fig. 2) homologous to PPDKs from prokaryotes [53% identity with Bacteroides symbiosus (29)], other protists [50% identity with G. lamblia (30) and 48% identity with E. histolytica (31)] and plants [45% identity with Flaveria trinervia (32)] (see Fig. 2 for consensus). The T. brucei PPDK homologue contains a specific insertion of 23 amino acids at position 295, and its last the C-terminal amino acids (AKL) conform to type 1 peroxisomal targeting signal, which has been shown to be effective also in targeting of proteins to glycosomes (33).
Figure 2.
Amino acid sequence of PPDK from T. brucei. The underlined 23-aa sequence is unique for the T. brucei PPDK, and the glycosomal targeting motif (AKL) located at the C-terminal end is bold faced and in italic. Identical amino acids and conservative amino acid substitutions between PPDK sequences from Bacteroides symbiosus (29), Entamoeba histolytica (31), Flaveria trinervia (32), and T. brucei are indicated by stars and open circles, respectively.
Expression of the PPDK homologue during the developmental cycle of T. brucei 427 was examined by Northern (data not shown) and Western blots (Fig. 5). Hybridization of a PPDK probe to equal amounts of bloodstream and procyclic forms RNA showed a much more abundant (about 10×) 4-kb mRNA in procyclic forms as compared with bloodstream forms (data not shown). This differential expression also was reflected at the protein level because Western blot analysis using mouse polyclonal antibodies to T. brucei recombinant PPDK revealed a 100-kDa protein in procyclic forms whereas no signal could be detected in the long–slender bloodstream form. However, the nondividing short–stumpy bloodstream form appeared to contain a very low amount of PPDK.
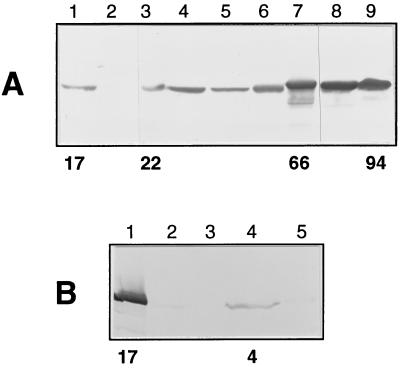
Figure 5.
Western blot analysis of trypanosomatids. In A, lysates of 107 cells from T. brucei 427 procyclic forms (lane 1) and bloodstream forms (lane 2), T. congolense procyclic forms (lane 3), epimastigote forms (lane 4), and bloodstream forms (lane 5), T. vivax bloodstream forms (lane 6), T. cruzi epimastigote forms (lane 7), Phytomonas (lane 8), and C. fasciculata choanomastigote forms (lane 9) were probed with mouse polyclonal anti-PPDK. In B, lysates of 3 × 107 cells from T. brucei 427 procyclic forms (lane 1), T. brucei GuTat short–stumpy form (lane 2) and long–slender form (lane 3), L. amazonensis promastigote forms (lane 4), and amastigote forms (lane 5) were analyzed as in A.
Functional Expression of PPDK in E. coli.
To test the function of the T. brucei-PPDK homologue, the PPDK gene containing a C-terminal polyhistidine tag was expressed in E. coli by using the pET vector system (Novagen) and was purified by Ni2+ nitriloacetic acid–agarose column. A 100-kDa protein recognized by anti-poly(his) (Novagen) and anti-T. brucei PPDK antibodies was purified to homogeneity with a specific activity of 2.5 units (μmole/min) per milligram of protein for catalysis of both the pyruvate and PEP formations. The divalent cation Mg2+ is required for activity whereas monovalent cations, K+ and NH4+, are not required for full enzymatic activity in either metabolic direction. For each of the six substrates involved in the forward (pyruvate production) or the reverse (PEP production) reactions, the affinity curves were hyperbolic with apparent Km values of 0.3 mM for pyruvate, 0.6 mM for ATP, 0.5 mM for inorganic phosphate, 40 μM for PEP, 7.5 μM for AMP and 50 μM for PPi. These values are similar to those obtained for other PPDKs (24, 34).
Subcellular Localization of PPDK in T. brucei.
Immunofluorescence analysis of procyclic forms with an anti-PPDK mAb (H121) showed a distinct punctate fluorescence characteristic of a glycosomal localization (Fig. 3B) as observed with a mAb specific for the glycosomal aldolase (Fig. 3A). No signal was observed with bloodstream forms (data not shown). To confirm this subcellular localization, the presence of PPDK was examined in sucrose-gradient purified glycosomes (Fig. 4). Western blot analysis revealed a 100-kDa protein recognized by anti-PPDK (Fig. 4B, lane 2) that comigrated with an abundant glycosomal protein (Fig. 4C, lane 2) described as a p90 protein specifically expressed in procyclic forms (35).
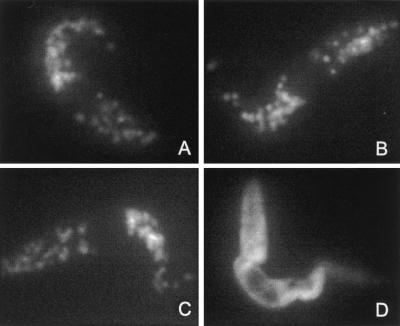
Figure 3.
Immunofluorescence analysis of T. brucei procyclic forms (EATRO 1125). Untransfected cells were stained with anti-aldolase (A) or anti-PPDK H121 (B) mAbs; anti-poly(His) was used to stain cells expressing histidine-tagged PPDK, which contains the AKL glycosomal targeting motif (PPDK34) (C) or histidine-tagged PPDK depleted of AKL glycosomal targeting motif (PPDK35) (D).
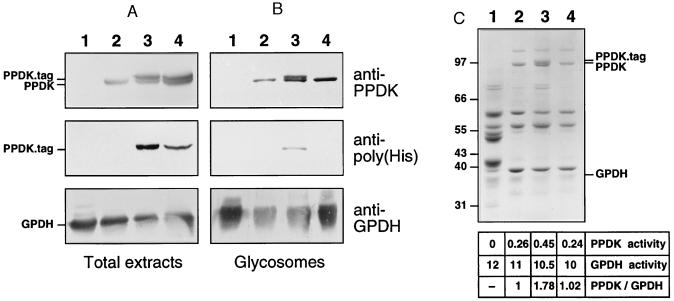
Figure 4.
Western blot analysis of T. brucei. Lysates of 107 cells (A) or 5 μg of glycosomal proteins (B) were analyzed with anti-PPDK, anti-poly(His), or anti-GPDH, and 20 μg of glycosomal proteins (C) were analyzed by Coomassie blue staining. Lanes: 1, bloodstream forms of T. brucei 427; 2, procyclic forms of T. brucei EATRO 1125; 3, procyclic forms of T. brucei EATRO 1125 expressing histidine-tagged PPDK, which contains AKL glycosomal targeting motif (PPDK34); 4, procyclic forms of T. brucei EATRO 1125 expressing histidine-tagged PPDK depleted for AKL glycosomal targeting motif (PPDK35). Antibodies used are indicated on the right margin of B, the position of tagged-PPDK, PPDK, and GPDH is indicated on the left margin of A and on the right margin of C, and the molecular weight markers are indicated on the left margin of C. The table in C indicates PPDK and GPDH activity as measured in glycosome preparations [milliunits (μg protein)−1] and the ratio PPDK activity/GPDH activity with an arbitrary value of 1 for EATRO-glycosomes.
To determine the role of the C-terminal AKL motif, which resembles a type 1 peroxisomal targeting signal, we used an epitope tag composed of six N-terminal histidine residues to mark unmodified (PPDK34) or AKL-deleted PPDK (PPDK35). The two recombinant PPDK genes were expressed in procyclic forms of T. brucei (EATRO1125 strain) and their subcellular localization analyzed by immunofluorescence with antibodies specific for the poly(His) (Novagen). The PPDK34 transfectant clearly showed a glycosomal localization (Fig. 3C), indicating that the N-terminal poly(His) epitope tag did not affect glycosomal targeting. In contrast, the PPDK35 transfectant presented a diffuse cytoplasmic staining (Fig. 3D). Western blot analysis on crude extracts of PPDK34 and PPDK35 transfectants revealed two high molecular weight proteins stained by anti-PPDK; the upper one, which was recognized by anti-poly(His), was recombinant PPDK (Fig. 4A). Identical results were obtained with glycosomes isolated from the PPDK34 transfectant whereas only endogenous PPDK was detected in glycosomes from the PPDK35 transfectant (Fig. 4B), which indicates that the C-terminal AKL motif was necessary for targeting PPDK toward glycosomes.
Coomassie blue staining revealed a single 100-kDa protein in glycosomes of untransfected cells and PPDK35 transfectant whereas a 100-kDa doublet with two bands of almost identical abundance was present in PPDK34 transfectant glycosomes (Fig. 4C). This profile was identical to the one obtained by Western blotting using anti-PPDK (Fig. 4B); the extra 100-kDa protein in PPDK34 transfectant was recognized by the anti-poly(His). This indicated that the endogenous PPDK, which was expressed at the same level as recombinant PPDKs, was the major 100-kDa protein present in glycosomes of procyclic forms. Native PPDK represents ≈5 to 10% of glycosomal proteins (Fig. 4C) and produced a yield of 0.26 units per milligram of glycosomal protein, indicating that the specific activity of PPDK is in the range of 2 units per milligram of enzyme.
Detection of PPDK Activity and Protein in Different Trypanosomatids.
PPDK activity was assayed in the forward reaction on crude extracts from bloodstream and procyclic forms of T. brucei strain 427. The level of PPDK activity in procyclic forms [17 milliunits (mg protein)−1] was identical to values obtained for other T. brucei strains (EATRO1125 and Stib247). In contrast, the high background level observed in bloodstream forms, mainly caused by a high PK activity that interfered with the PPDK assay, precluded the determination of PPDK activity on crude extracts. PPDK activity could be assayed on glycosomal fractions devoid of PK activity, which is only present in the cytosolic fraction. Glycosomes from procyclic forms (EATRO1125) yielded 0.26 units (mg protein)−1 whereas no activity was detected in those purified from bloodstream forms (strain 427) (Fig. 4C), indicating that PPDK activity was only present in procyclic forms. Noteworthy, PPDK activity was 15 times enriched in glycosomal fractions, as observed for GPDH activity (data not shown).
The presence of PPDK in other trypanosomatids such as T. congolense, T. vivax, T. cruzi, Phytomonas, C. fasciculata, and L. amazonensis was assayed by Western blotting by using either polyclonal anti-PPDK antibodies (Fig. 5) or mAb H122 (data not shown). Every trypanosomatid analyzed so far contained a 100-kDa protein that crossreacted with anti-PPDK antibodies; the apparent molecular weight of the African trypanosome PPDKs was slightly smaller as compared with other parasites. PPDK was present in T. congolense (procyclic, epimastigote, and bloodstream forms), T. vivax (bloodstream forms), and epimastigote forms (Fig. 5) and trypomastigotes forms (data not shown) of T. cruzi. A very faint signal was observed in intracellular forms (amastigote) of L. amazonensis; the signal was more intense in the insect forms (promastigote). The most intense signal was observed in plant and insect parasites (Phytomonas and C. fasciculata). The level of PPDK activity, as assayed on crude extracts of different trypanosomatids, was comparable with the signal intensity obtained by Western blotting (Fig. 5). The insect form of L. amazonensis presented the lowest detectable amount [4 milliunits (mg protein)−1], and that of C. fasciculata presented the highest [94 milliunits (mg protein)−1].
DISCUSSION
Here, we reported the cloning of a single copy gene encoding PPDK of T. brucei, which was expressed functionally in E. coli. Several lines of evidence show that PPDK is a glycosomal protein: (i) Although T. brucei PPDK shares a high level of identity with all homologues characterized so far, it distinguishes itself by a much higher calculated isoelectric point (8.25 vs. 5.20 to 6.51), which is a common feature of glycosomal proteins (36); (ii) immunofluorescence analysis with anti-PPDK displays the distinct punctate fluorescence characteristic of glycosomal localization; (iii) PPDK and glycosomal GPDH activities are both enriched ≈15 times in glycosomal fractions, and Western blot analysis indicates that both proteins also are enriched in these fractions; (iv) immunofluorescence and Western blot analyses of procyclic forms transfected with recombinant PPDK showed that the AKL motif present at the C-terminal extremity of the enzyme, which resembles the canonical type 1 peroxisome targeting motif (33), is necessary for PPDK import into the glycosomes; and (v) glycosomal fractions of procyclic forms expressing recombinant AKL-bearing PPDK contain about twice as much PPDK protein and PPDK activity as compared with procyclic forms transfected with recombinant PPDK lacking the AKL motif or untransfected cells (Fig. 4C). Anti-PPDK antibodies recognize, in procyclic forms of T. brucei, a 100-kDa glycosomal protein that comigrates with one of the most abundant glycosomal proteins, both being absent from glycosomes of long–slender bloodstream form. In addition, epitope-tagged recombinant PPDK and the 100-kDa glycosomal protein, which are expressed at the same level in the PPDK34 transfectant, are stained equally with anti-PPDK antibodies. This indicates that PPDK and the 100-kDa glycosomal protein previously called p90 (35), which represent ≈10% of glycosomal proteins, are the same protein.
PPDK is regulated developmentally in T. brucei; procyclic forms contain ≈10× more protein than the short–stumpy bloodstream form whereas the protein is absent from crude extracts or glycosomal fractions of the long–slender bloodstream form. The presence of a low level of PPDK in the short–stumpy bloodstream form could reflect another aspect of the preadaptation of the parasite’s carbohydrate (4) and mitochondrial (37, 38, 39) metabolisms to their subsequent differentiation into procyclic forms. Furthermore, Northern blot analysis (data not shown) shows clearly that the long–slender form contains PPDK encoding mRNA (≈10× less than procyclic forms), but no protein is detectable. This indicates that regulation of expression partly occurs at the post-transcriptional level, as observed for most of the rare, nuclear-encoded mitochondrial cytochrome subunits analyzed so far (38).
The native PPDK purified with glycosomes from T. brucei and the recombinant T. brucei PPDK purified to homogeneity from E. coli yield about the same specific activity (≈2 units per milligram of enzyme). This low specific activity also was observed for recombinant or native PPDKs isolated from other organisms (24, 25, 34, 40) as well as for the other PPi-dependent glycolytic enzymes (9, 41).
PPDK and the few other pyrophosphate-dependent enzymes involved in glycolysis (PPi-phosphofructokinase, PEP carboxyphosphotransferase) were described in prokaryotes, plants, and protists but never in vertebrates (8, 9, 10). These enzymes are frequent in protists, as shown by their increasing list: Trichomonadidae (Trichomonas), Diplomonadina (Giardia), Euglenophyta, Amobida (Entamoeba and Naegleria), Apicomplexa (Toxoplasma), anaerobic ciliates (Trimyema and Isotrichia), and now the trypanosomatids. The most primitive protists (Giardia, Trichomonas and Entamoeba), which live in anaerobiosis, developed a PPi-dependent glycolysis, which suggests that they exploit the advantage that PPi-dependent glycolysis is more efficient than conventional glycolysis (up to five vs. two ATP molecules produced per glucose molecule) (41). The precise role of PPi-dependent enzymes detected in the aerobically grown protists is not clear. The well characterized glycolysis of trypanosomatids appears to be conventional, with, as expected, no indication of a direct involvement of PPi (4, 6, 42). Furthermore, PPDK is the only PPi-dependent enzyme characterized so far in trypanosomatids. Interestingly, glycosomal ATP-dependent phosphofructokinase of T. brucei shares the highest degree of identity with PPi-dependent phosphofructokinases, suggesting that a PPi-dependent enzyme present in trypanosomatid ancestors evolved recently into an ATP-dependent enzyme (43). This observation suggests that the PPi-dependent enzymes and metabolism used by some prokaryotes and primitive eukaryotes were replaced by ATP-dependent enzymes and metabolic processes in most of the present-day organisms, but phylogenetic analysis of ATP-PFK and PPi-PFK displays more contrasted results (10).
In B. symbiosus, PPDK appears to fulfill the glycolytic role (forward reaction) normally assigned to PK whereas the enzyme from the C4-plants and Propionibacterium shermanii functions in the gluconeogenic direction (reverse reaction) (41). The recent detection of a PK activity in Giardia (11) raised questions about the PPDK function in the amitochondriate protists. In T. brucei, which contains a cytosolic PK (6), we showed that PPDK is a glycosomal protein, but the question remains, In which direction does the enzyme work under physiological conditions, and what is its role in the intermediate metabolism of trypanosomatids? The presence of both PK and PPDK activities have been reported only in Giardia (11) and the bacteria P. shermanii (41), where they are located in the cytosol. In T. brucei, these two enzymes are present in separate subcellular compartments, consistent with their possible simultaneous involvement in different metabolic processes. To be a glycolytic enzyme, PPDK would need pyrophosphate, which might be present in glycosomes because the organelle is devoid of pyrophosphatase activity (43) and PPi is produced by enzymes such as the glycosomal hypoxanthine-guanine phosphoribosyltransferase involved in the purine salvage pathway (6). Every PPDK analyzed so far, including the enzyme from T. brucei, B. symbiosus (forward reaction), and plants (reverse reaction), presents a much higher Km for pyruvate, ATP, and inorganic phosphate than for PEP, ADP, and PPi. These observations suggest that the glycosomal PPDK from T. brucei potentially can function in both forward and reverse directions. The metabolic constraints and the glycosomal substrate concentrations should determine the direction in which the reaction proceeds in vivo.
All adaptative forms of trypanosomatids produce succinate during glucose metabolism, indicating a mitochondrial activity, except the long–slender bloodstream form of T. brucei, which is devoid of any mitochondrial activity involved in carbohydrate metabolism (4, 7). The latter is also the only trypanosomatid analyzed so far that lacks PPDK. Thus, it is tempting to suggest that PPDK is associated indirectly with a mitochondrial function in trypanosomatids. Bloodstream forms of T. brucei secrete all of the pyruvate produced aerobically whereas, in other trypanosomatids, a major part of PEP is redirected to the glycosome for the formation of l-malate, which is further reduced to succinate through a reversed C4 part of the mitochondrial Krebs cycle. A possible role for PPDK could be the production, in glycosomes, of PEP from cytosolic pyruvate, which would be further reduced into l-malate. A glycosomal enzyme required to produce l-malate from PEP (PEP carboxykinase) was detected in every trypanosomatid analyzed to date, excepted the bloodstream forms of T. brucei. Noteworthy, the chloroplastic PPDK, which provides PEP for CO2 fixation, functions this way. PPDK also could be the first step of a glycosomal gluconeogenesis. Obviously, we are far from understanding the physiological role of the PPDK in trypanosomatids. Further experiments need to be performed to answer this question. Most importantly, the viability of trypanosomes depleted of the PPDK gene needs to be determined because the absence of PPi-dependent enzymes in vertebrates enables use of phosphonate analogues as potential drugs for trypanosomatids as well as for other parasites (9).
Acknowledgments
We are grateful to L. Vanhamme, D. Salmon, and E. Pays, who provided us with the pTSA-3′proc expressing vector and with useful help about parasite transfections, and F. R. Opperdoes and P. Michels for useful advice about glycosome preparations and helpful discussions. We would like to thank N. Bakalara, C. Bourget, A. Cuvillier, P. Minoprio, and C. Vedrenne for supplying us with trypanosomatids, D. Parzy for DNA sequencing, and M. Müller, P. Michels, and G. Merlin for critical reading of the manuscript. This work was supported by the CNRS, the Ministère de l’Enseignement Supérieur et de la Recherche, the Groupement de Recherche: Centre National de le Recherche Scientifique/ Direction Générale des Armées, and the Pôle Aquitaine Santé (Médicament).
ABBREVIATIONS
- PPDK
pyruvate, phosphate dikinase
- PEP
phosphoenolpyruvate
- PFK
phosphofructokinase
- GPDH
glycerol-3-phosphate dehydrogenase
- PK
pyruvate kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF048689).
References
- 1.Cavalier-Smith T. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marché S, Roth C, Philippe H, Dollet M, Baltz T. Mol Biochem Parasitol. 1995;71:15–26. doi: 10.1016/0166-6851(95)00029-z. [DOI] [PubMed] [Google Scholar]
- 3.Jukes J, Jirku M, Dolezel D, Kral’ova I, Hollar L, Maslov D A. J Mol Evol. 1997;44:521–527. doi: 10.1007/pl00006176. [DOI] [PubMed] [Google Scholar]
- 4.Fairlamb A H, Opperdoes F R. In: Carbohydrate Metabolism in Cultured Cells. Morgan M J, editor. New York: Plenum; 1986. pp. 183–224. [Google Scholar]
- 5.Vickerman K. Br Med Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 6.Opperdoes F R. Annu Rev Microbiol. 1987;41:127–151. doi: 10.1146/annurev.mi.41.100187.001015. [DOI] [PubMed] [Google Scholar]
- 7.Bowman I B R, Flynn I W. In: Biology of the Kinetoplastida. Lumsden W H R, Evans D A, editors. London: Academic; 1976. pp. 436–458. [Google Scholar]
- 8.Coombs G H, Müller M. In: Biochemistry and Molecular Biology of Parasites. Marr J J, Müller M, editors. London: Academic; 1995. pp. 33–47. [Google Scholar]
- 9.Mertens E. Parasitol Today. 1993;9:122–126. doi: 10.1016/0169-4758(93)90169-g. [DOI] [PubMed] [Google Scholar]
- 10.Müller M. In: Evolutionary Relationships Among Protozoa. Combs G H, Vickerman K, Sleigh M A, Warren A, editors. London: Chapman & Hall; 1998. pp. 109–132. [Google Scholar]
- 11.Park J-H, Schofield P J, Edwards M R. Exp Parasitol. 1997;87:153–156. doi: 10.1006/expr.1997.4206. [DOI] [PubMed] [Google Scholar]
- 12.Jack R M, Black S J, Reed S L, Davis C E. Infect Immun. 1984;43:445–448. doi: 10.1128/iai.43.1.445-448.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanham S M, Godfrey D G. Exp Parasitol. 1970;28:521–534. doi: 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- 14.Brun R, Schönenberger M. Acta Trop. 1979;36:289. [PubMed] [Google Scholar]
- 15.Gray M A, Ross C A, Taylor A M, Luckins A G. Acta Trop. 1984;42:99–111. [PubMed] [Google Scholar]
- 16.Taylor A E R, Baker J R. In Vitro Methods for Parasite Cultivation. London: Academic; 1978. [Google Scholar]
- 17.Opperdoes F R, Borst P, Spits H. Eur J Biochem. 1977;76:21–28. doi: 10.1111/j.1432-1033.1977.tb11566.x. [DOI] [PubMed] [Google Scholar]
- 18.Toner J J, Weber M M. Biochem Biophys Res Commun. 1972;46:652–660. doi: 10.1016/s0006-291x(72)80190-6. [DOI] [PubMed] [Google Scholar]
- 19.Bringaud F, Baltz T. Mol Biochem Parasitol. 1992;52:111–122. doi: 10.1016/0166-6851(92)90040-q. [DOI] [PubMed] [Google Scholar]
- 20.Campbell D A. Nucleic Acids Res. 1989;17:458. doi: 10.1093/nar/17.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bates P F, Swift R A. Gene. 1983;26:137–146. doi: 10.1016/0378-1119(83)90183-x. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Saiki R K, Gelfang D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Herlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 24.South D J, Reeves R E. Methods Enzymol. 1975;42:187–191. doi: 10.1016/0076-6879(75)42114-0. [DOI] [PubMed] [Google Scholar]
- 25.Benzamin M. Methods Enzymol. 1975;42:192–199. doi: 10.1016/0076-6879(75)42115-2. [DOI] [PubMed] [Google Scholar]
- 26.Misset O, Opperdoes F R. Eur J Biochem. 1984;144:475–483. doi: 10.1111/j.1432-1033.1984.tb08490.x. [DOI] [PubMed] [Google Scholar]
- 27.Fazekas de St. Groth S, Scheidegger D. J Immunol Methods. 1980;35:1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 29.Pocalyko D J, Carroll L J, Martin B M, Babbitt P C, Dunaway-Mariano D. Biochemistry. 1990;29:10757–10765. doi: 10.1021/bi00500a006. [DOI] [PubMed] [Google Scholar]
- 30.Nevalainen L, Hrdy I, Müller M. Mol Biochem Parasitol. 1996;77:217–223. doi: 10.1016/0166-6851(96)02604-7. [DOI] [PubMed] [Google Scholar]
- 31.Bruchhaus I, Tannich E. Mol Biochem Parasitol. 1993;62:153–156. doi: 10.1016/0166-6851(93)90193-2. [DOI] [PubMed] [Google Scholar]
- 32.Rosche E, Westhoff P. FEBS Lett. 1990;273:116–121. doi: 10.1016/0014-5793(90)81064-u. [DOI] [PubMed] [Google Scholar]
- 33.Clayton C, Häusler T, Blattner J. Microbiol Rev. 1995;59:325–344. doi: 10.1128/mr.59.3.325-344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatch M D, Slack C R. Methods Enzymol. 1975;42:212–219. [Google Scholar]
- 35.Parsons M, Nielsen B. Exp Parasitol. 1990;70:276–285. doi: 10.1016/0014-4894(90)90109-p. [DOI] [PubMed] [Google Scholar]
- 36.Wierenga R K, Swinkels B, Michels P A M, Osinga K, Misset O, Van Beeumen J, Gibson W C, Postma J P M, Borst P, Opperdoes F R, et al. EMBO J. 1987;6:215–221. doi: 10.1002/j.1460-2075.1987.tb04741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feagin J E, Jasmer D P, Stuart K. Mol Biochem Parasitol. 1986;20:207–214. doi: 10.1016/0166-6851(86)90100-3. [DOI] [PubMed] [Google Scholar]
- 38.Priest J W, Hajduk S L. J Bioenerg Biomembr. 1994;26:179–192. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- 39.Vickerman K. Nature (London) 1965;208:762–766. doi: 10.1038/208762a0. [DOI] [PubMed] [Google Scholar]
- 40.Chastain C J, Thompson B J, Chollet R. Phytosynth Res. 1996;49:83–89. doi: 10.1007/BF00029430. [DOI] [PubMed] [Google Scholar]
- 41.Wood H G, O’Brien W E, Michaels G. Adv Enzymol Relat Areas Mol Biol. 1977;45:85–155. doi: 10.1002/9780470122907.ch2. [DOI] [PubMed] [Google Scholar]
- 42.Cazzulo J J. FASEB J. 1992;6:3153–3161. doi: 10.1096/fasebj.6.13.1397837. [DOI] [PubMed] [Google Scholar]
- 43.Michels P A M, Chevalier N, Opperdoes F R, Rider M, Rigden D J. Eur J Biochem. 1997;250:698–704. doi: 10.1111/j.1432-1033.1997.00698.x. [DOI] [PubMed] [Google Scholar]