Abstract
The signal recognition particle (SRP) of eukaryotic cells is a cytoplasmic ribonucleoprotein machine that arrests the translational elongation of nascent secretory and membrane proteins and facilitates their transport into the endoplasmic reticulum. The spatial pathway of SRP RNA processing and ribonucleoprotein assembly in the cell is not known. In the present investigation, microinjection of fluorescently tagged SRP RNA into the nucleus of mammalian cells was used to examine its intranuclear sites of localization. Microinjection of SRP RNA into the nuclei of normal rat kidney (NRK) epithelial cells maintained at 37°C on the microscope stage resulted in a very rapid initial localization in nucleoli, followed by a progressive decline of nucleolar signal and an increase of fluorescence at discrete sites in the cytoplasm. Nuclear microinjection of a molecule corresponding to a major portion of the Alu domain of SRP RNA revealed a pattern of rapid nucleolar localization followed by cytoplasmic appearance of signal that was similar to the results obtained with full-length SRP RNA. In contrast, a molecule corresponding to the S domain of SRP RNA did not display nucleolar localization to the extent observed with full-length SRP RNA. An SRP RNA molecule lacking helix 6 of the S domain displayed normal nucleolar localization, whereas one lacking helix 8 of the S domain did not. These results, obtained by direct, real-time observation of fluorescent RNA molecules inside the nucleus of living mammalian cells, suggest that the processing of SRP RNA or its ribonucleoprotein assembly into the SRP involves a nucleolar phase.
The signal recognition particle (SRP) is a ribonucleoprotein machine that delivers certain nascent polypeptides to specific recognition components on the cytoplasmic face of the endoplasmic reticulum membrane for translocation of secretory or membrane proteins (1, 2). The RNA component of the SRP contains two elements related to the human and rodent Alu families of interspersed repetitive DNA sequences connected by a unique sequence, the S domain (3–5). SRP RNA (Fig. 1) has an overall secondary structure that has been highly conserved during evolution of the Bacteria, Archaea, and Eukarya (6–9). SRP RNA associates with six proteins termed, in mammalian cells, SRP72, SRP68, SRP54, SRP19, SRP14, and SRP9 (10). The regions of SRP RNA bound by protein originally were identified by mild micrococcal nuclease digestion, which results in cleavage of the SRP into two subparticles containing the Alu and S domains, respectively (11). The Alu domain, which consists of approximately 100 nucleotides at the 5′ end of SRP RNA base-paired with approximately 50 nucleotides at the 3′ end, together with its associated proteins SRP9 and SRP14, comprises the translational arrest activity of SRP (12). The S domain consists of approximately 150 nucleotides of core SRP RNA sequence situated between the two Alu sequences and is bound by the remaining four proteins SRP19, SRP54, SRP68, and SRP72 (2). It is within the S domain of SRP that nascent polypeptide signal sequence recognition mediated by SRP54 and the protein translocation activity mediated by SRP68 and SRP72 reside (13–17).
Figure 1.
Human SRP RNA. Regions denoted by the numerals 2–8 follow the nomenclature of Larsen and Zwieb (6); helix 1 is present only in archaebacterial SRP RNA. The two arrows denote sites at which the SRP is cleaved into two subparticles by mild micrococcal nuclease digestion (11).
Purified SRP can be disassembled into its constituents (SRP RNA, SRP19, SRP54, and the two heterodimeric protein complexes SRP9/14 and SRP68/72) and subsequently reassembled from these purified components into a functional particle (18). Subsequent studies using SRP proteins expressed in vitro from their cloned cDNAs have demonstrated a stepwise pathway of SRP assembly. Previous association of SRP19 with SRP RNA is required for SRP54 assembly (18), possibly directed by an SRP19-induced conformational change in SRP RNA (19–24). The SRP9 and SRP14 proteins first associate with each other in the absence of SRP RNA, forming an SRP9/14 heterodimer, which then assembles onto SRP RNA (25–27). Although SRP68 and SRP72 are disassembled from purified SRP as a heterodimer (18), SRP68 and SRP72 associate inefficiently with each other in the absence of SRP RNA (28), suggesting that previous formation of the SRP68/72 heterodimer does not occur. Instead SRP68, which by itself binds SRP RNA weakly, appears to serve as an adapter for the assembly of SRP72, which does not bind SRP RNA in the absence of SRP68 (28).
Although these in vitro studies have revealed considerable insight into the RNA–protein and protein–protein interactions involved in SRP assembly, the intracellular site or sites of SRP biosynthesis are not known. A previous investigation demonstrated that at least partial assembly of the SRP occurs in the nucleus in Xenopus oocytes and implicated the Alu domain of SRP RNA in nuclear export (29). SRP RNA (previously termed 7SL RNA) is an RNA polymerase III transcript bearing a triphosphate 5′ end (3). Although recovered primarily in cytoplasmic fractions, some SRP RNA also has been observed in nuclear fractions from mammalian cells (30, 31), and a portion of the nuclear SRP RNA was noted to fractionate with purified nucleoli (31).
In the present investigation we have investigated the intranuclear localization of SRP RNA in mammalian cells by nuclear microinjection of fluorescent RNA. This method affords direct, real-time observation of the movements of wild-type and mutant RNAs in living cells (32–37). Our results demonstrate that fluorescent SRP RNA microinjected into the nucleus of mammalian cells rapidly localizes in the nucleolus. In addition, the use of mutant SRP RNAs has revealed that the Alu domain and helix 8 of SRP RNA are required for nucleolar localization.
MATERIALS AND METHODS
Preparation of Fluorescent RNAs.
Canine SRP RNA was transcribed from XbaI-digested plasmid pSP7SL (ref. 7; provided by Steven Ogg and Peter Walter, University of California, San Francisco) using T7 RNA polymerase (GIBCO/BRL). Human SRP RNA and the mutant human SRP RNAs ΔH6, ΔH8, and Δ35 were transcribed from previously described plasmids (ref. 21; kindly provided by Christian Zwieb, University of Texas Health Center at Tyler) digested with DraI (phR, pΔH6 and pΔH8) or BamHI (pΔ35). The SRP Alu domain RNA ΔL21, consisting of nucleotides 1–68 linked to nucleotides 277–301 of human SRP RNA, was transcribed from DraI-digested plasmid pΔL21 (C. Zwieb, personal communication). Human pre-tRNAser was transcribed from AvaI-digested plasmid pUC19pSer (38). All RNAs were transcribed in the presence of 5-(3-aminoallyl)-UTP (35, 36) using T7 RNA polymerase. The aminoallyl-U substituted RNA was recovered by ethanol precipitation, coupled to tetramethylrhodamine-6-isothiocyanate, purified, and microinjected into the nucleus of normal rat kidney (NRK) cells as previously described (32–36).
Cell Culture and RNA Microinjection.
NRK epithelial cells were cultured in F-12K medium (JRH Biosciences, Lenexa, KS) containing 10% fetal bovine serum, 50 μg/ml of streptomycin, and 50 μg/ml of penicillin. Cells were plated on glass coverslips 36–48 hr before the subsequent period of observation (35). Nuclear microinjections were performed as previously described (32, 33, 35).
RESULTS AND DISCUSSION
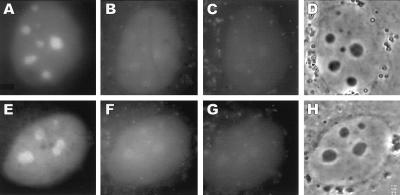
To investigate the intranuclear localization of SRP RNA in a dynamic context in living cells, we used the method of fluorescent RNA cytochemistry developed in our laboratory (32–37). As shown in Fig. 2, fluorescent SRP RNA microinjected into the nucleus of NRK cells became rapidly localized in nucleoli. Although the earliest timepoints after nuclear microinjection shown in Fig. 2 are 3 and 6 min (A and E, respectively), in other experiments extensive nucleolar localization of SRP RNA was observed within 30 sec after nucleus microinjection (data not shown). Twenty to 30 min after nuclear microinjection of SRP RNA the nucleolar fluorescence decreased and signal appeared in the cytoplasm (Fig. 2 B and F), localized in foci, which became even more prominent ≈60 min after nucleus microinjection (Fig. 2 C and G). We have not further characterized the cytoplasmic localization of fluorescent SRP RNA, but the results shown in Fig. 2 conform to the expected nucleus-to-cytoplasm pathway of SRP RNA. No nucleolar localization was observed after nuclear microinjection of a fluorescent pre-tRNA (Fig. 3). Moreover, there was no nucleolar localization of U6 small nuclear RNA or of intron-containing or intron-lacking pre-mRNAs in our previous studies (32, 34). Thus, the observed nucleolar localization of fluorescent SRP RNA is specific.
Figure 2.
Rapid nucleolar localization of fluorescent SRP RNA after microinjection into the nucleus. Rhodamine-labeled canine SRP RNA was prepared as detailed in Materials and Methods and microinjected into the nucleus of NRK cells. Two representative experiments are shown. Fluorescence at 3 min (A), 26 min (B), and 66 min (C) after nuclear microinjection. Note the early, transient localization in nucleoli (A), followed by appearance in the cytoplasm (B and C). (D) Phase-contrast micrograph taken at 66 min. Fluorescence at 6 min (E), 33 min (F), and 56 min (G). Note again the early, transient nucleolar localization (E) and subsequent cytoplasmic appearance (F and G). (H) Phase-contrast image taken at 56 min. Results similar to those shown were obtained with fluorescent human SRP RNA.
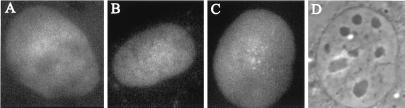
Figure 3.
Pre-tRNA controls. A human seryl transfer RNA precursor was rhodamine-labeled and injected into the nucleus of NRK cells. Three representative cells are shown. (A) 3 min after microinjection. (B and C) 4 min after microinjection. (D) Phase-contrast micrograph of the cell in C.
A major advantage of fluorescent RNA microinjection in the study of intracellular RNA dynamics is that it affords the analysis of sequence elements required or dispensable for proper traffic and localization (32–35, 37). We therefore used mutants of SRP RNA to investigate sequences important for nucleolar localization. As noted above, the SRP is a rather extreme case of a bipartite ribonucleoprotein particle in that its protein components are clustered in two relatively confined domains of the RNA’s overall folded structure with a less or noncovered region of RNA in between (Fig. 1). This was first suggested by mild micrococcal nuclease digestion experiments that cleaved the SRP into two subparticles without apparent loss of protein (11) and subsequently has been confirmed by studies of RNA chemical modification in the SRP (39) and refined reassembly experiments (2). We therefore decided to first use SRP mutants that represent one or the other of the two known ribonucleoprotein domains of the SRP (Fig. 4).
Figure 4.
Mutant SRP RNAs used in this investigation. ΔH6, ΔH8, and Δ35 were described previously (21); ΔL21 was constructed as detailed in Materials and Methods and was kindly provided by C. Zwieb. ΔL21 consists of the region within the Alu domain that is the binding site for SRP9 and SRP14 proteins. Δ35 corresponds to the S domain. ΔH6 is the complete SRP RNA lacking only helix 6, and ΔH8 is the complete SRP RNA lacking only helix 8.
When a mutant SRP RNA corresponding to a portion of the Alu domain (ΔL21 in Fig. 4) was microinjected into the nucleus of NRK cells it became localized in nucleoli within 1–3 min (Fig. 5 B and C), which is temporally and spatially similar to the results obtained with wild-type SRP RNA (Fig. 2). At later times after microinjection (16 min, Fig. 5D) the Alu domain RNA displayed a reduced nucleolar level and appeared in the cytoplasm in a patched pattern, again similar to the results obtained with wild-type SRP RNA (Fig. 2).
Figure 5.
Nuclear microinjection of fluorescent SRP RNA mutant corresponding to the SRP9/14 binding portion of the Alu domain (ΔL21 in Fig. 4). (A) Phase-contrast image taken 1 min after microinjection. Fluorescence at 1 min (B), 3 min (C), and 16 min (D) after microinjection. (E and F) Phase-contrast and fluorescence of a noninjected cell. Note the same early, transient nucleolar localization as observed with full-length SRP RNA in Fig. 2.
Different results were observed when an RNA corresponding to the S domain of SRP RNA (Δ35 in Fig. 4) was microinjected into the nucleus. In some cases, as illustrated in the example shown in Fig. 6 A–D, there was no evidence of nucleolar localization whatsoever. In other instances, as in the example shown in Fig. 6 E–I, there was only a very brief (30 sec) initial nucleolar localization that disappeared by 3 min (Fig. 6F). The basis of this variation from one cell to another in these experiments with the S domain mutant is not presently understood, and both types of results are shown in Fig. 6 to accurately convey the findings. The main point, however, is that in no case did the S domain mutant display as pronounced a nucleolar localization as either the wild-type SRP RNA (Fig. 2) or the Alu domain mutant (Fig. 5).
Figure 6.
SRP RNA mutant corresponding to the S domain (Δ35 in Fig. 4). Two representative experiments are shown, illustrating the extremely short-lived nucleolar localization with this S domain mutant. (A) 1 min after microinjection. (B) 3 min. (C) 24 min. (D) Phase-contrast at 24 min. Note that in contrast to wild-type SRP RNA (Fig. 2) or the Alu domain mutant (Fig. 4), which show nucleolar localization for at least 1–3 min after nuclear microinjection, the S domain mutant displays no nucleolar localization at 1 min (A). The pattern is a punctate nucleoplasmic distribution. (E–H) In this second experiment, nucleolar localization was observed at 30 sec after nuclear microinjection (E) but did not persist. (F) 3 min. (G) 10 min. (H) 19 min. (I) Phase-contrast at 19 min.
The results obtained for Δ35 (Fig. 6) reflect the nucleolar localization of the S domain itself, in the absence of the Alu domain. We therefore went on and investigated the importance of S domain elements in the context of otherwise intact (i.e., Alu domain-containing) SRP RNA. Accordingly, we used SRP RNA mutants lacking only helix 6 or helix 8 in the S domain (ΔH6 and ΔH8 in Fig. 4). When microinjected into the nucleus, SRP RNA lacking helix 6 of the S domain displayed a pattern very similar to that observed for wild-type SRP RNA and the Alu domain RNA: it became localized in nucleoli within 30 sec and remained in the nucleoli for at least another 2.5 min (Fig. 7 A and C). At subsequent times, the nucleolar signal decreased but, unlike the results obtained for wild-type SRP RNA and the Alu domain RNA, there was no evidence of cytoplasmic appearance of the helix 6-lacking SRP RNA (Fig. 7 D–F). In contrast, when an SRP RNA lacking helix 8 of the S domain was microinjected into the nucleus, it displayed little initial nucleolar localization altogether (Fig. 8 A and C) and displayed no appearance in the cytoplasm. The fact that the S domain mutant, Δ35, displayed very weak, if any, nucleolar localization (Fig. 6) whereas helix 6-lacking mutant (containing the Alu domain) localized in nucleoli to the same extent as wild-type SRP RNA suggests that the Alu domain is the major nucleolar localization factor. According to this interpretation, when the Alu domain is absent, the S domain is incapable of nucleolar localization. When the Alu domain is present as a dominant nucleolar localization signal, an additional requirement of helix 8 in the S domain becomes apparent as a secondary factor.
Figure 7.
SRP RNA mutant lacking helix 6 (ΔH6 in Fig. 4). Shown are fluorescence patterns at 30 sec (A), 3 min (C), 12 min (D), 26 min (E), and 43 min (F) after nuclear microinjection. (B) Phase-contrast at 1 min. Note the early, transient nucleolar localization, similar to that observed for full-length SRP RNA (Fig. 2) and the Alu domain mutant (Fig. 5).
Figure 8.
SRP RNA mutant lacking helix 8 (ΔH8 in Fig. 4). Shown are the fluorescence patterns at 30 sec (A), 3 min (C), 13 min (D), 25 min (E), and 43 min (F). (B) Phase-contrast at 1 min.
We have attempted to detect nuclear sites of endogenous SRP RNA by in situ hybridization but our results so far have not been consistent (M.R.J., J. C. Politz, S. Kilroy, and T.P., unpublished results). SRP RNA has a secondary structure in which there are no single-stranded regions longer than eight nucleotides (loop of helix 4 in Fig. 1). SRP RNA therefore may be a relatively intractable in situ hybridization target. Other investigators have commented on the failure of immobilized complementary oligonucleotides to select SRP RNA from mammalian cell extracts (40), consistent with the notion that the secondary structure and ribonucleoprotein structure of SRP RNA constitutes an unreactive entity for complementary oligonucleotides. The fact that we have detected reproducible, sequence-specific SRP RNA in situ hybridization signals in the cytoplasm of NRK (and other mammalian) cells is consistent with the possibility that nucleolar SRP RNA has a particularly protected ribonucleoprotein conformation or deeply buried nucleolar location. Finally, if the nucleolus is indeed a site of SRP RNA processing or SRP assembly, it would be anticipated that the steady-state nucleolar content of SRP RNA would be very low relative to the cytoplasmic level, perhaps below the detection limit of in situ hybridization. It is noteworthy that SRP RNA previously was detected in RNA extracted from highly purified rat hepatoma cell nucleoli (31). More recently, a percentage of SRP RNA has been observed to cofractionate with HeLa cell nucleoli (James Mitchell and Kathleen Collins, University of California, Berkeley, personal communication).
The analysis of the regions of SRP RNA important for nucleolar localization presented here obviously constitutes only an initial undertaking, and we have not dissected the molecule in detail. The Alu domain and helix 8 of SRP RNA implicated in nucleolar localization in this investigation do not display any obvious sequence homology to other small nucleolar RNAs such as U3 or U8 (41, 42), nor to identified nucleolar localization elements in small nucleolar RNAs such as RNase mitochondrial RNA processing RNA (33), RNase P RNA (34), or intron-derived small nucleolar RNAs (43, 44). Thus, it appears that there are multiple RNA sequence zip codes for nucleolar localization. Like other previously described RNA localization signals, the nucleolar address elements in SRP RNA may operate either as RNA sequences per se or as distinctive ribonucleoprotein signatures assembled on specific RNA domains.
The fact that the nucleolar association of SRP RNA observed in this investigation is short-lived is not compatible with a nonspecific binding phenomenon. The introduced SRP RNA has an initial affinity for the nucleolus, possibly as the result of previous events in the nucleoplasm, but then apparently is released from the nucleolar structure. The fact that the Alu domain and helix 6-lacking SRP RNA mutants also display this transient rather than a permanent nucleolar localization adds weight to the notion that this observed nucleolar association-dissociation phenomenon is a reflection of a normal nucleolar transit of SRP RNA. It is to be noted that microinjection of other fluorescent small nucleolar RNAs into the nucleus of mammalian cells leads to a more long-lived nucleolar localization, as has been observed with RNase mitochondrial RNA processing RNA for example (33), again indicating that the transient nucleolar association of SRP RNA observed in the present investigation is likely to have a functional basis.
The rapidity with which SRP RNA (and the nucleolus-localizing SRP RNA mutants) reach the nucleolus from their nucleoplasmic sites of microinjection is striking, occurring extensively within only a minute at 37°C in these living cell experiments. We do not know if this rapid nucleolar localization reflects a mediated transport process or simple diffusion, nor is the necessary information about the intranuclear physical chemical milieu available to make this distinction. In a recent investigation we found that oligodeoxynucleotides (smaller than the SRP RNA studied here) move within the nucleus of living mammalian cells with mean translational diffusion times similar to those measured in aqueous solution (45). This finding suggests that, despite an apparently crowded intranuclear environment (46), there is free volume inside the nucleus in which molecular motion is driven solely by thermal forces, i.e., diffusion.
SRP RNA is relatively unique among eukaryotic RNAs in having no modified nucleotides in those organisms in which this has been examined (47). As an RNA polymerase III transcript (48), the processing of SRP RNA likely consists simply of the removal of one or few 3′-terminal uridylate residues encoded by the CTT(T) pol III terminator, as well as the recently discovered posttranscriptional addition of uridylate or adenylate nucleotides or formation of a 2′,3′-cyclic phosphate on the 3′-terminal U residue of SRP RNA (49, 50). The results of the present investigation raise the possibility that these SRP RNA 3′ events may occur in the nucleolus. The observed nucleolar localization of microinjected SRP RNA also may reflect the possibility that its partial or complete assembly into the SRP ribonucleoprotein particle is obligatorily linked to the nucleolus in some way. The nucleolar affinity of SRP RNA may be related to a transient ribonucleoprotein assembly stage or conformation that subsequently is modified, by further assembly, into a particle having an attenuated nucleolar affinity, thereby prompting its release and export to the cytoplasm. In this connection it is of interest to note that switches in the conformation of SRP RNA have been described as a function of the binding of specific SRP proteins (23, 24, 39, 51). Our results raise the possibility that SRP RNA (as partially or completely assembled SRP) may be exported from the nucleus in association with one of the ribosomal subunits. This possibility is compatible with the fact that the functional cycle of the signal recognition particle in the cytoplasm involves its interaction not only with the N terminus of nascent polypeptides but also with the ribosome itself (2, 52). Finally, the finding that SRP RNA associates with the nucleolus may have evolutionary implications. A large number of recent findings, in addition to the present ones, suggest that the nucleolus may be plurifunctional, facilitating numerous aspects of RNA processing, ribonucleoprotein assembly, and nuclear export in addition to its well-defined role in ribosome synthesis (53).
Acknowledgments
We gratefully acknowledge the essential help of Christian Zwieb (University of Texas Health Science Center), who kindly provided the wild-type and mutant human SRP RNA plasmids. We thank Steven Ogg and Peter Walter (University of California, San Francisco) for providing the canine SRP RNA plasmid, Cecilia Guerrier-Takada and Sidney Altman (Yale University) for the human pre-tRNA plasmid, and Kathleen Collins (University of California-Berkeley) for communicating unpublished results. We are grateful to Susan Kilroy for expert assistance in transcription, fluorescent labeling, and purification of RNA. This investigation was supported by National Institutes of Health Grant GM-21595-21 to T.P. and funds awarded to the Worcester Foundation for the study of RNA by the Harold G. and Y. Leila Mathers Foundation.
ABBREVIATIONS
- SRP
signal recognition particle
- NRK
normal rat kidney
References
- 1.Walter P, Lingappa V R. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- 2.Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 3.Li W-Y, Reddy R, Henning D, Epstein P, Busch H. J Biol Chem. 1982;257:5136–5142. [PubMed] [Google Scholar]
- 4.Ullu E, Murphy S, Melli M. Cell. 1982;29:195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Blobel G. Nature (London) 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 6.Larsen N, Zwieb C. Nucleic Acids Res. 1991;19:209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strub K, Moss J, Walter P. Mol Cell Biol. 1991;11:3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Althoff S, Selinger D, Wise J A. Nucleic Acids Res. 1994;22:1933–1947. doi: 10.1093/nar/22.11.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwieb C, Larsen N. Nucleic Acids Res. 1997;25:107–108. doi: 10.1093/nar/25.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter P, Blobel G. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundelfinger E D, Krause E, Melli M, Dobberstein B. Nucleic Acids Res. 1983;11:7362–7374. doi: 10.1093/nar/11.21.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel V, Walter P. Nature (London) 1986;320:81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- 13.Krieg U C, Walter P, Johnson A E. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurzchalia T V, Wiedmann M, Girschovich A S, Bochkareva E S, Bielka H, Rapoport T A. Nature (London) 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 15.Wiedmann M, Kurzchalia T V, Bielka H, Rapoport T A. J Cell Biol. 1987;104:201–208. doi: 10.1083/jcb.104.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel V, Walter P. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- 17.Siegel V, Walter P. Proc Natl Acad Sci USA. 1988;85:1801–1805. doi: 10.1073/pnas.85.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter P, Blobel G. Cell. 1983;34:525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- 19.Römisch K, Webb J, Lingelbach K, Gausepohl H, Dobberstein B. J Cell Biol. 1990;111:1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwieb C. Nucleic Acids Res. 1985;13:6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwieb C. Nucleic Acids Res. 1991;19:2955–2960. doi: 10.1093/nar/19.11.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwieb C, Ullu E. Nucleic Acids Res. 1986;14:4639–4657. doi: 10.1093/nar/14.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker K P, Black S D, Zwieb C. Biochemistry. 1995;34:11989–11997. doi: 10.1021/bi00037a041. [DOI] [PubMed] [Google Scholar]
- 24.Gowda K, Zwieb C. Nucleic Acids Res. 1997;25:2835–2840. doi: 10.1093/nar/25.14.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strub K, Walter P. Mol Cell Biol. 1990;10:777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu K, Chang D-Y, Maraia R J. J Biol Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- 27.Chang D-Y, Newitt J A, Hsu K, Bernstein H D, Maraia R J. Nucleic Acids Res. 1997;25:1117–1122. doi: 10.1093/nar/25.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lütcke H, Prehn S, Ashford A J, Remus M, Frank R, Dobberstein B. J Cell Biol. 1993;121:977–985. doi: 10.1083/jcb.121.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X-P, Bataille N, Fried H M. J Cell Sci. 1994;107:903–912. doi: 10.1242/jcs.107.4.903. [DOI] [PubMed] [Google Scholar]
- 30.Gurney T, Eliceiri G L. J Cell Biol. 1980;87:398–403. doi: 10.1083/jcb.87.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy R, Li W-Y, Henning D, Choi Y C, Nohga K, Busch H. J Biol Chem. 1981;256:8452–8457. [PubMed] [Google Scholar]
- 32.Wang J, Cao L-G, Wang Y-L, Pederson T. Proc Natl Acad Sci USA. 1991;88:7391–7395. doi: 10.1073/pnas.88.16.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson M R, Cao L-G, Wang Y-L, Pederson T. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobson M R, Cao L-G, Taneja K, Singer R H, Wang Y-L, Pederson T. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson M R, Pederson T. In: Analysis of mRNA Formation and Function: Methods in Molecular Genetics. Richter J D, editor. New York: Academic; 1997. pp. 341–359. [Google Scholar]
- 36.Jacobson M R, Pederson T, Wang Y-L. In: Cell Biology: A Laboratory Handbook. Celis J E, editor. IV. New York: Academic; 1998. pp. 5–10. [Google Scholar]
- 37.Jacobson M R, Pederson T. Nucleic Acids Res. 1998;26:756–760. doi: 10.1093/nar/26.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mamula M J, Baer M, Craft J, Altman S. Proc Natl Acad Sci USA. 1989;86:8717–8721. doi: 10.1073/pnas.86.22.8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreazzoli M, Gerbi S A. EMBO J. 1991;10:767–777. doi: 10.1002/j.1460-2075.1991.tb08008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matera A G, Frey M R, Margelot K, Wolin S L. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy R, Henning D, Busch H. J Biol Chem. 1979;254:11097–11105. [PubMed] [Google Scholar]
- 42.Reddy R, Henning D, Busch H. J Biol Chem. 1985;260:10930–10935. [PubMed] [Google Scholar]
- 43.Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 44.Bachellerie J P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 45.Politz J C, Browne E S, Wolf D E, Pederson T. Proc Natl Acad Sci USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pederson T. J Mol Biol. 1998;227:147–159. doi: 10.1006/jmbi.1997.1618. [DOI] [PubMed] [Google Scholar]
- 47.Reddy, R. (1998) Nucleic Acids Rev. 16, Suppl., r71–r85. [DOI] [PMC free article] [PubMed]
- 48.Zieve G, Benecke B-J, Penman S. Biochemistry. 1977;16:4520–4525. doi: 10.1021/bi00639a029. [DOI] [PubMed] [Google Scholar]
- 49.Gu J, Shumyatsky G, Makan N, Reddy R. J Biol Chem. 1997;272:21989–21993. doi: 10.1074/jbc.272.35.21989. [DOI] [PubMed] [Google Scholar]
- 50.Sinha M, Gu J, Chen Y, Reddy R. J Biol Chem. 1998;273:6853–6859. doi: 10.1074/jbc.273.12.6853. [DOI] [PubMed] [Google Scholar]
- 51.Thomas Y, Bui N, Strub K. Nucleic Acids Res. 1997;25:1920–1929. doi: 10.1093/nar/25.10.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogg S C, Walter P. Cell. 1995;81:1075–1084. doi: 10.1016/s0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]
- 53.Pederson, T. (1998) Nucleic Acids Res. 26, in press. [DOI] [PMC free article] [PubMed]