Abstract
Signal transductions by the dual-function CXCR4 and CCR5 chemokine receptors/HIV type 1 (HIV-1) coreceptors were electrophysiologically monitored in Xenopus laevis oocytes that also coexpressed the viral receptor CD4 and a G protein-coupled inward-rectifying K+ channel (Kir 3.1). Large Kir 3.1-dependent currents generated in response to the corresponding chemokines (SDF-1α for CXCR4 and MIP-1α; MIP-1β and RANTES for CCR5) were blocked by pertussis toxin, suggesting involvement of inhibitory guanine nucleotide-binding proteins. Prolonged exposures to chemokines caused substantial but incomplete desensitization of responses with time constants of 5–7 min and recovery time constants of 12–19 min. CXCR4 and CCR5 exhibited heterologous desensitization in this oocyte system, suggesting possible inhibition of a common downstream step in their signaling pathways. In contrast to chemokines, perfusion with monomeric or oligomeric preparations of the glycoprotein of Mr 120,000 (gp120) derived from several isolates of HIV-1 did not activate signaling by CXCR4 or CCR5 regardless of CD4 coexpression. However, adsorption of the gp120 from a T-cell-tropic virus resulted in CD4-dependent antagonism of CXCR4 response to SDF-1α, whereas gp120 from macrophage-tropic viruses caused CD4-dependent antagonism of CCR5 response to MIP-1α. These antagonisms could be partially overcome by high concentrations of chemokines and were specific for coreceptors of the corresponding HIV-1 isolates, suggesting that they resulted from direct interactions of gp120–CD4 complexes with coreceptors and that they did not involve the desensitization pathway. These results indicate that monomeric or oligomeric gp120s specifically antagonize CXCR4 and CCR5 signaling in response to chemokines, but they do not exclude the possibility that gp120s might also function as weak agonists in some cells. The gp120-mediated disruption of CXCR4 and CCR5 signaling may contribute to AIDS pathogenesis.
Fusion of the viral membrane with the cell surface membrane during infection by HIV type 1 (HIV-1) involves collaboration between the CD4 receptor and a coreceptor (1–6). Coreceptors are seven transmembrane G protein-coupled receptors for proinflammatory chemokines. The major coreceptor for macrophage-tropic (M-tropic) HIV-1 isolates is CCR5, which is activated by the chemokines MIP-1α, MIP-1β, and RANTES, whereas that for T-cell-tropic HIV-1 isolates is CXCR4, which is activated by SDF-1 (7). The gp120 envelope glycoprotein (glycoprotein of apparent Mr 120,000) of HIV-1 forms a ternary complex with CD4 and coreceptors that displaces 125I-labeled chemokines (8–13).
The roles of chemokine receptor signaling in HIV-1 infection remain uncertain. Several mutations in CCR5 and CXCR4 that prevent G protein activation do not perturb their coreceptor activities (14, 15), suggesting that virus-induced activation of these chemokine receptors cannot be essential for infection. Nevertheless, evidence reported during preparation of this manuscript indicated that monomeric or oligomeric gp120s derived from several M-tropic isolates of HIV-1 could induce CCR5-mediated Ca2+ mobilization and chemotaxis in a proportion of activated CD4-positive human T lymphocytes (16). However, similar glycoprotein preparations from other immunodeficiency viruses that also use CCR5 failed to activate signaling. Moreover, gp120 preparations from T-cell-tropic viruses did not activate signaling by CXCR4. These results imply that gp120 shed from nests of cells infected with some M-tropic HIV-1 might chemoattract uninfected CD4- and CCR5-positive cells or enhance their susceptibilities to infection or to apoptotic stimuli. Another recent report indicated that gp120s from various T-cell-tropic or M-tropic HIV-1 isolates did not induce detectable Ca2+ mobilization in activated CD4-positive T lymphocytes, despite the substantial responses of these same cells to chemokines (17). However, this group detected significant gp120-mediated increases in tyrosine phosphorylation of Pyk2 tyrosine kinase and presented evidence that this response depended on CXCR4 and CCR5. The implications of these results are discussed below.
A potential complication in this field derives from evidence that gp120 binding to CD4 in T lymphocytes can activate the associated Lck tyrosine kinase, with resultant activation of the Raf-mitogen-activated protein kinase pathway and of transcription factors including NF-κB (18–20). In lymphocytes and other cells, tyrosine kinase-signaling events can cause G protein-independent Ca2+ mobilization with resultant Pyk2 activation (21, 22) or chemotaxis (23). Moreover, signal transduction by chemokine receptors also activates src family tyrosine kinases and mitogen-activated protein kinases (22, 24, 25). Because of the likelihood of overlap and cross-talk between the gp120-activated Lck and chemokine receptor-signaling pathways, thorough understanding of gp120 signaling will ultimately require controlled studies of cells that lack either or both of these pathways. In addition, the assays such as Ca2+ mobilization that have been commonly used to study chemokine receptor signaling generally appear to have high backgrounds and poor signal-to-noise ratios. As one approach toward analyzing these issues, we developed a sensitive system in which chemokine receptor signaling in Xenopus oocytes is coupled to a G protein-activated inward-rectifying K+ channel (Kir 3.1; refs. 26 and 27). Kir 3.1 is a high conductivity channel that is promiscuously activated by Gβγ-subunits (28–31). Xenopus oocytes lack CD4, Lck, and chemokine receptors but contain many heterotrimeric G proteins and have been widely used to study chemokine receptors and other G protein-coupled receptors (32–34). Our results suggest that this system may be exceptionally useful for quantitatively and directly monitoring G protein activation by CXCR4 and CCR5. In this system, gp120s were CD4-dependent and tropism-specific antagonists of CXCR4 and CCR5 responses to chemokines.
MATERIALS AND METHODS
Reagents and cDNA Clones.
Chemokines were from PeproTech (Rocky Hill, NJ) and pertussis toxin was from Calbiochem. Monomeric T-cell-tropic gp120 (IIIB) was donated by Shiu-lok Hu (Bristol-Myers Squibb). Monomeric M-tropic gp120 Ba-L was a gift from Ray Sweet (SmithKline Beecham) and oligomeric JR-FL gp120 in a complex with the extracellular domain of gp41 was provided by James Arthos and Anthony Fauci (National Institute of Allergy and Infectious Diseases, Bethesda). Full-length SDF-1 was provided by Ian Clark-Lewis (University of British Columbia, Vancouver). Vectors and cDNA clones were gifts of the following investigators; Kir 3.1, Henry Lester (California Institute of Technology, Pasadena, CA); mGluR 2, Shigetada Nakanishi (Kyoto University); pBF expression vector, John Adelman (The Vollum Institute); CXCR4 cDNA, Frank R. Jirik (University of British Columbia, Vancouver); CCR5 cDNA, John P. Moore (Aaron Diamond Research Institute, New York). CD4 polyclonal antibody was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease, and was contributed by Michael Phelan.
125I-gp120 Binding to Xenopus Oocytes and HeLa Cells.
Samples (10–25 μg) of gp120 were labeled with [125I]Bolton-Hunter reagent (ICN), and aliquots were adsorbed onto oocytes in a frog Ringer’s solution 96 mM NaCl/2 mM KCl/1.8 mM CaCl2/1 mM MgCl2/5 mM Hepes (pH 7.5) for 1–2 h at room temperature with gentle shaking. Each oocyte was then separately washed for 5 min in Ringer’s solution and counted in a gamma counter. Binding of a constant trace amount of 125I-gp120 onto HeLa and HeLa-CD4 cells (clone H1-J) (35) was done in the presence of various concentrations of unlabeled gp120 for times indicated at 37°C.
Oocyte Expression.
Stage V–VI oocytes were collected from anesthetized Xenopus laevis and defolliculated with collagenase (Boehringer Mannheim). Oocytes were incubated at 16°C in frog Ringer’s solution supplemented with 2.5 mM sodium pyruvate (Sigma), 0.5 mM theophylline (Sigma), and 50 μg/ml gentamycin (Life Technologies, Grand Island, NY). cDNAs were subcloned into oocyte expression vectors (pOG-1 or pBF) at a site between 5′- and 3′-untranslated Xenopus β-globin sequences (36). Linearized plasmids were transcribed in vitro with T7 or SP6 polymerase, and 5–50 ng of capped cRNA were microinjected into oocytes on the day of harvest. Membranes were isolated from oocytes and western immunoblotting was done with a sheep anti-human CD4 antiserum or a rabbit antiserum to CCR5 (37).
Electrophysiology.
Electrophysiological recording was done 2–5 d after cRNA injection. Two-electrode voltage clamp was performed with a GeneClamp 500 amplifier interfaced to a Digidata 1200 A/D (Axon Instruments, Foster City, CA). The interface was controlled with an IBM-compatible computer running pclamp, version 6.0 (Axon Instruments, Foster City, CA). Microelectrodes were filled with 3 M KCl and had tip resistances of 0.1–1.5 MΩ. The oocytes were placed in a small chamber continually perfused with high K+ Ringer’s solution [100 mM KCl/2 mM NaCl/1.8 mM CaCl2/1 mM MgCl2/5 mM Hepes (pH 7.5)]. Agonists and blockers were applied by bath perfusion. The holding potential was set at −30 mV and current–voltage records were obtained during 250-ms voltage jumps to potentials between +40 and −100 mV. Desensitization kinetics were determined by least squares fit to single exponential functions. Kinetics of recovery from desensitization were determined by measuring the responses in different oocytes at specific intervals. The responses were then normalized to the peak response and fitted to an exponential function. Where indicated, oocytes were incubated with 1 μg/ml pertussis toxin in Ringer’s solution for 48 h before recording.
RESULTS
Chemokine-Induced Activation of Kir 3.1 in Xenopus Oocytes.
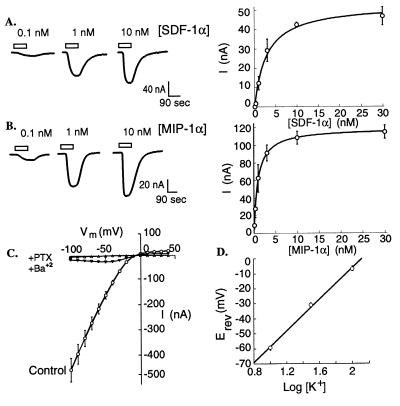
Capped cRNAs for chemokine receptors and Kir 3.1 were coinjected into oocytes, and voltage clamp current recordings were obtained 2–3 d after injection. Oocytes were clamped at −30 mV in high K+–Ringer’s solution, and varying concentrations of receptor agonists were applied by bath perfusion. Because glutamate is known to activate Kir 3.1 through binding to the G protein-coupled metabotropic glutamate receptor mGluR 2 (34), we used this receptor as a control. This response saturated at glutamate concentrations of 50–100 μM with half-maximal stimulation at 5.96 ± 1.43 μM (n = 4; results not shown). As shown in Fig. 1A, the inward currents induced by SDF-1α in oocytes expressing CXCR4 were also concentration dependent. Saturation was reached between 10 and 30 nM SDF-1α with a half-maximal stimulation at 2.84 ± 0.57 nM (n = 3). Similarly, MIP-1α activated CCR5 (Fig. 1B) with half-maximal activation at 0.94 ± 0.04 nM MIP-1α (n = 3). In addition, the CCR5 agonists MIP-1β and RANTES activated currents in oocytes expressing this receptor with half-maximal stimulations at 0.33 ± 0.04 nM (n = 3) and 4.0 ± 0.65 nM (n = 3), respectively. The activities of these agonists were specific, because concentrations of 10 nM MIP-1α did not induce currents in oocytes coexpressing Kir 3.1 and CXCR4 (n = 2). Conversely, 10 nM SDF-1α did not induce currents in oocytes that coexpressed Kir 3.1 and CCR5 (n = 7). Control uninjected oocytes and oocytes lacking chemokine receptors or Kir 3.1 did not show any currents when the ligands were applied (data not shown). Protein immunoblots confirmed the presence of CCR5 in membranes of oocytes that had been injected with this cRNA (data not shown).
Figure 1.
Left traces: Activation of currents in oocytes coexpressing Kir 3.1 with CXCR4 (A) and CCR5 (B). Inward currents were seen in response to superfusion of ligands for the duration indicated by the bar above the trace. The holding potential was −30 mV, and the recording solutions were K+–Ringer’s solution containing 100 mM KCl. Right plots: Concentration dependence of currents. Data were fitted to a rectangular hyperbola, yielding the following EC50 values for each ligand: SDF-1α, 2.8 ± 0.6 nM (n = 3); MIP-1α, 0.9 ± 0.04 (n = 3). (C) Inhibition of CXCR4/Kir 3.1-mediated currents by pertussis toxin (PTX) pretreatment (1 μg/ml, 48 h) or BaCl2 (100 μM). Each point represents mean ± SEM, n = 4. (D) Reversal potential (Erev) of currents produced by 3 nM SDF-1α in oocytes coexpressing Kir 3.1 and CXCR4 depends on external K+ concentrations. Line shown is least squares fit (54 mV/decade) through points representing mean ± SEM (n = 3).
To verify that the currents induced by chemokine application resulted from activation of the G protein-dependent K+ channel Kir 3.1, we characterized the properties of these currents. The voltage dependencies of currents induced by activation of mGluR 2, CXCR4, and CCR5 showed strong inward rectification at all agonist concentrations, as expected for Kir 3.1 (33). This is illustrated in Fig. 1C, which shows the current–voltage curve for CXCR4 activation by 1 nM SDF-1α. These data also show strong inhibition of these currents by the K+ channel blocker Ba2+ and their abolishment by pertussis toxin. In addition, the reversal potentials of currents induced by 3 nM SDF-1α changed by 54.2 mV per 10-fold change in extracellular K+, close to the prediction of the Nernst equation for a K-selective conductance (Fig. 1D). These data show that the currents induced in oocytes by chemokine applications resulted from activation of Kir 3.1 by specific G protein receptor-signaling pathways.
Desensitization of Chemokine Receptor Signaling.
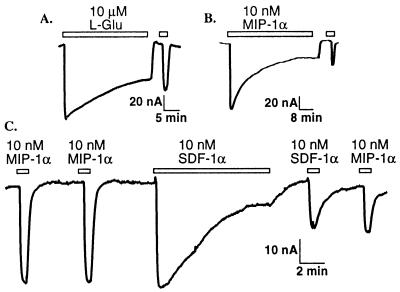
The currents induced by 10 μM l-Glu in oocytes that coexpressed mGluR 2 and Kir 3.1 are further analyzed in Fig. 2A. Glutamate-dependent currents desensitized during prolonged agonist application with a time constant of 7.5 ± 1.2 min (n = 5), to a steady-state value representing 44.3 ± 5.1% of the initial response. Fig. 2B shows a response of the CCR5 receptor to a prolonged application of 10 nM MIP-1α. The response decayed with a time constant of 5.3 ± 0.4 min (n = 11) to 46.7 ± 6.0% of the initial response. Similarly, prolonged application of 10 nM SDF-1α resulted in attenuation of the CXCR4 response to 23.2 ± 2.7% of the initial value with a time constant of 6.1 ± 0.6 min (n = 10) (Fig. 2C). Following washout of agonists, the responses recovered slowly from desensitization. The time constant for recovery of CXCR4 following steady-state desensitization induced by SDF-1α was 12.8 ± 1.7 min, and the time constant for CCR5 recovery after desensitization with MIP-1α was approximately 18.7 ± 4.4 min.
Figure 2.
Desensitization of chemokine and glutamate receptors. (A) Decay of inward current during continued application of 10 μM l-Glu (indicated by bar above trace) and partial recovery following washout of agonist in a representative oocyte coexpressing mGluR 2 and Kir 3.1. (B) Similar homologous desensitization of CCR5 chemokine receptor response during long exposure to MIP-1α. (C) Heterologous desensitization of CCR5 response by prolonged CXCR4 activation in oocytes coinjected with CXCR4, CCR5, and Kir 3.1 cRNA.
Cross-desensitization in oocytes coexpressing CXCR4 and CCR5 together with Kir 3.1 was also apparent. Fig. 2C shows that brief nondesensitizing applications of MIP-1α induced similar current amplitudes. Following a prolonged exposure to SDF-1α that resulted in desensitization of the CXCR4 response, the CCR5 response to MIP-1α was also desensitized. This cross-desensitization by SDF-1α in oocytes coexpressing CXCR4, CCR5, and Kir 3.1 resulted in inhibition of CCR5 responses to MIP-1α by 50.2 ± 5.9% (n = 6). Conversely, desensitization with MIP-1α inhibited SDF-1α activation of CXCR4 by 66.0 ± 9.5% (n = 4). Similarly, desensitization with SDF-1α in oocytes that coexpressed CXCR4, mGluR 2, and Kir 3.1 inhibited l-Glu activation of mGluR 2 by 23.8 ± 5.5% (n = 4), whereas desensitization with l-Glu inhibited SDF-1α activation of CXCR4 by 84 ± 5.9% (n = 3). This heterologous desensitization suggests the involvement of effectors downstream of receptor activation including G proteins and/or ion channels.
Effects of HIV-1 gp120 on Signal Transduction by CXCR4 and CCR5.
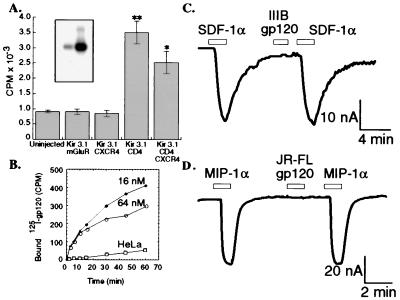
The gp120s used in this investigation were highly purified (e.g., see Fig. 3A Inset) and were active in binding to oocytes that had been injected with CD4 cRNA (Fig. 3A) as well as to human HeLa–CD4 cells but not to control HeLa cells (Fig. 3B). The latter data also show the kinetics at which different concentrations of monomeric gp120 saturate CD4 on surfaces of Hela–CD4 cells. Abundant expression of CD4 in oocyte membranes was also demonstrated by protein immunoblotting (results not shown). To determine whether gp120 induces a signal when it binds to CD4 and the coreceptor, we tested highly purified T-cell-tropic gp120 IIIB as well as M-tropic oligomeric gp120 JR-FL and monomeric gp120 Ba-L in the K+ channel activation assay. As shown in Fig. 3C, no currents were generated in response to perfusion of HIV-1 gp120 IIIB (16 nM) onto voltage-clamped oocytes coexpressing CD4, CXCR4, and Kir 3.1. This was substantiated with a total of 39 oocytes from 14 batches. Moreover, the currents induced by SDF-1α did not decrease after short applications (30–90 s) of gp120 IIIB. Similarly, M-tropic oligomeric gp120 from the JR-FL isolate of HIV-1 was perfused onto oocytes that coexpressed CD4, CCR5, and Kir 3.1. An inward current was induced when 10 nM MIP-1α was applied, whereas M-tropic gp120 (20 nM) did not elicit any currents in the same oocyte, and brief applications of JR-FL gp120 did not decrease subsequent MIP-1α activation of CCR5 (Fig. 3D). Identical results were obtained in 12 oocytes from three different batches. In addition, the monomeric M-tropic Ba-L gp120 also failed to induce signals in oocytes that had been coinjected with cRNAs for CD4, CCR5, and Kir 3.1 (n = 3). Superfusion of gp120 onto voltage-clamped oocytes for longer times (10–20 min) was similarly ineffective at inducing currents in oocytes that were coinjected with cRNAs that encode CD4, respective coreceptors, and Kir 3.1 (data not shown).
Figure 3.
(A) 125I-gp120 IIIB binding to Xenopus oocytes. 125I-gp120 IIIB (1.2 × 106 cpm/ml) was used for binding to one batch of oocytes that coexpressed combinations of Kir 3.1 with mGluR 2, CD4, and CXCR4 as well as to uninjected control oocytes (**P < 0.0001; *P < 0.0024 by the Student’s unpaired t test). Each column represents mean ± SEM for five determinations. (Inset) 125I-gp120 was analyzed by gel electrophoresis followed by autoradiography. Different amounts were loaded onto the two lanes. (B) 125I-gp120 IIIB binding to HeLa–CD4 (clone H1-J) and to control HeLa cells. Cell cultures were incubated at 37°C with 125I-gp120 IIIB for various times in the presence of unlabeled gp120 IIIB at a concentration of 16 nM (•) or 64 nM (○). Nonspecific background was determined by incubating HeLa cells lacking CD4 with the same amount of 125I-gp120 IIIB in the absence of unlabeled gp120 IIIB (□). C and D demonstrate that HIV-1 gp120 IIIB and JR-FL do not induce currents in oocytes coexpressing CD4 and Kir 3.1 with their respective coreceptors. (C) An inward current was produced by application of 3 nM SDF-1α to a representative oocyte coexpressing CD4, CXCR4, and Kir 3.1. Application of 16 nM monomeric gp120 IIIB did not induce a current and also did not reduce the subsequent SDF-1α activation of CXCR4. (K+–Ringer’s solution, membrane potential = −30 mV). (D) Similar experiment showing lack of effect of JR-FL gp120 in a representative oocyte coexpressing CD4, CCR5, and Kir 3.1. An inward current was observed during application of 10 nM MIP-1α, but brief application of 20 nM oligomeric JR-FL gp120 did not induce a current or reduce the magnitude of the subsequent response to MIP-1α.
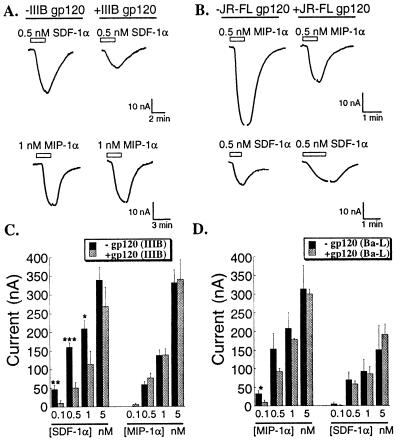
We then examined effects of prolonged preincubations with high concentrations of T-cell-tropic as well as M-tropic gp120s on chemokine receptor signaling. Saturating concentrations of highly purified gp120 IIIB were incubated for 1–2 h with oocytes that coexpressed CD4, CXCR4, CCR5, and Kir 3.1 before voltage clamping. Similar to results with short applications, preadsorption of gp120 IIIB did not activate a significant baseline K+ current [slope conductances at −95 mV were 6.3 ± 0.5 (n = 20) and 6.9 ± 0.3 (n = 19) for control and gp120 treated oocytes, respectively]. However, this preincubation with T-cell-tropic gp120 IIIB significantly decreased SDF-1α-induced currents compared with untreated control oocytes (Fig. 4A, top tracings; P < 0.02; n = 30), without affecting MIP-1α-induced currents (Fig. 4A, bottom tracings). Similarly, oocytes that were preincubated for 2 h with a saturating concentration of oligomeric M-tropic JR-FL gp120 had significantly decreased responses to MIP-1α compared with the untreated control oocytes (Fig. 4B, top right; P < 0.02; n = 4), whereas the SDF-1α-induced currents in the same oocytes did not change (Fig. 4B, bottom tracings). These inhibitory effects were completely dependent on CD4 expression in the oocytes (results not shown).
Figure 4.
Extensive adsorption of monomeric T-cell-tropic or oligomeric M-tropic gp120s specifically and competitively antagonize CXCR4 and CCR5 responses to chemokines in a tropism-specific manner. All oocytes coexpressed CXCR4, CCR5, CD4, and Kir 3.1. (A) Current records for two representative oocytes that were preincubated with 0.4 μM gp120 IIIB for 1–2 h before voltage clamping and superfusing with SDF-1α and MIP-1α at the concentrations indicated. (B) Oocytes were similarly preincubated with 0.3 μM oligomeric JR-FL gp120 for 1–2 h before analysis. (C) Monomeric gp120 IIIB inhibition of CXCR4 activation by SDF-1α over a range of concentrations, without any effect on CCR5 responses to different concentrations of MIP-1α in the same oocytes. The oocytes were preincubated with gp120 IIIB as in A, and measurements were recorded at −80 mV during voltage pulses (***P < 0.0002; **P < 0.0012; *P < 0.05; each column represents mean ± SEM; n = 5–6). (D) Monomeric gp120 Ba-L inhibition of CCR5 responses to different concentrations of MIP-1α, without any significant effect on CXCR4 responses to different concentrations of SDF-1α. The oocytes were preincubated with gp120 Ba-L as described for A, and measurements were recorded at −80 mV during voltage pulses (*P < 0.08; each column represents mean ± SEM; n = 3).
Similar analyses were then done over wide ranges of chemokine concentrations. As shown in Fig. 4C, extensive preadsorption with gp120 IIIB significantly decreased SDF-1α activation of CXCR4 at all concentrations compared with the control oocytes. However, this inhibition of signaling was proportionately greater at lower concentrations of SDF-1α. Specifically, the inhibition was 80% at 0.1 nM SDF-1α and approximately 20% at 5 nM SDF-1α. In contrast, extensive preadsorption of gp120 IIIB had no significant effect on CCR5 responses to MIP-1α in these same oocytes. These results confirm the specificity of the gp120 IIIB inhibition and suggest that a competitive mechanism may be involved in the inhibition of CXCR4 activation by SDF-1α. A similar analysis was also done with monomeric gp120 from M-tropic Ba-L isolate, with nearly identical results (see Fig. 4D). In this case, the inhibition was specific for CCR5 responses to MIP-1α and was greatest when the concentration of MIP-1α was low. The Ba-L gp120 had no significant effect on CXCR4-signaling responses to SDF-1α in these same oocytes.
DISCUSSION
Chemokine Receptor Signaling.
In oocytes that coexpressed CXCR4 and CCR5 with the G protein-coupled K+ channel Kir 3.1, channel activations were saturable functions of the appropriate chemokines (Fig. 1A and B). These currents were completely blocked by pertussis toxin (Fig. 1C), consistent with previous evidence for chemokine receptor coupling to Gi proteins (38, 39). The properties of the conductance activated by these ligands, including its K selectivity, voltage-dependence, and sensitivity to block by Ba2+ ions, are in accord with the properties of Kir 3.1 (26, 27). In agreement with this conclusion, oocytes lacking Kir 3.1 did not show detectable currents. Because opening of Kir 3.1 channels is mediated by direct interactions with Gβγ-subunits that are released when heterotrimeric G proteins are activated (28–31), this assay reflects primary G protein activation and does not depend on complex downstream signaling events such as Ca2+ fluxes or protein kinase cascades. Accordingly, the response of the system is highly reproducible, making it exceptionally useful for quantitative studies and for examining modulations caused by gp120 glycoproteins of immunodeficiency viruses.
Current responses desensitized in the continuous presence of chemokine with time constants of 5–8 min and reached steady-state levels that appeared receptor specific (e.g., see Fig. 2). Moreover, when CXCR4, CCR5, and mGluR 2 were coexpressed, heterologous desensitization was observed, as has been observed previously for coexpressed μ-opioid and serotonin-1A receptors in oocytes (32). Heterologous desensitization implies that the oocyte down-modulatory mechanism is induced by activation of all of these receptors. This mechanism may promiscuously inhibit all G protein-coupled receptors or it may inhibit a postreceptor step that is common to their signaling pathways. In contrast, exposure of mammalian cells to a chemokine causes rapid down-modulation and cessation of its signal within approximately 1–2 min (40, 41), without any desensitization of heterologous receptors (16). In that case, activation of chemokine receptors results in their rapid phosphorylation, binding of arrestins, and specific endocytosis (40–44).
Effects of gp120 on Signaling by CXCR4 and CCR5.
In contrast to the currents induced by chemokines in oocytes that coexpress CD4 and Kir 3.1 with CXCR4 or CCR5, brief superfusions with relatively high concentrations of highly purified oligomeric or monomeric gp120 derived from T-cell-tropic and M-tropic HIV-1 did not induce detectable signals in these same oocytes (see Fig. 3C and D). Signaling was also absent when applications of gp120 were increased up to 20 min (results not shown). Nevertheless, it was conceivable that gp120-induced receptor activation might occur more slowly than the time scale of these experiments. This might occur as a consequence of the relatively slow saturation of CD4 with gp120 as demonstrated in Fig. 3B and/or to a slow recruitment of coreceptors into ternary complexes. Clearly, however, our gp120 preparations were competent for binding to CD4 (Fig. 1) and for CD4-dependent interactions with specific coreceptors (Fig. 4). Moreover, the oligomeric JR-FL gp120 preparation was generously donated by James Arthos and Anthony Fauci, who recently reported that it induced rapid CCR5-mediated Ca2+ mobilization in activated CD4-positive human T lymphocytes (16). Consistent with previous reports (8, 9, 11–13), our gp120 preparations also caused CD4-dependent reductions in specific 125I-labeled chemokine binding to mammalian cells (S.L.K and D.K., unpublished data).
To address the absence of gp120-induced coreceptor activations in the oocyte superfusion assay, we preadsorbed oligomeric or monomeric gp120s for 1–2 h before assaying responses to chemokines. The baseline K+ currents were unaffected by the adsorptions, confirming an absence of chemokine receptor activation. Interestingly, however, extensive preadsorption of gp120 from the T-cell-tropic HIV-1 isolate IIIB strongly inhibited subsequent response of CXCR4 to SDF-1α, whereas extensive preadsorption of oligomeric or monomeric gp120 preparations from the M-tropic HIV-1 strains JR-FL and Ba-L inhibited CCR5-mediated signaling (see Fig. 4). These gp120-induced inhibitions were CD4 dependent and completely specific for the coreceptors that mediate infections by these isolates of HIV-1. This specificity clearly differs from activation-induced desensitization of signaling that inhibits bystander G protein-coupled receptors (see Fig. 2), suggesting instead a direct effect. In agreement with this interpretation, the gp120 IIIB-mediated inhibition of CXCR4 signaling was partially overcome at high concentrations of SDF-1α (see Fig. 4C), implying that it involves a competitive mechanism, and corresponding results were obtained for the gp120 Ba-L inhibition of CCR5 responses to MIP-1α (Fig. 4D). Together, these observations suggest that gp120–CD4 complexes bind directly to the appropriate chemokine receptors to competitively inhibit their activations by chemokines. This interpretation is concordant with previous evidence that gp120–CD4 complexes bind to chemokine receptors (10) and partially displace 125I-labeled chemokines (8, 9, 12, 13). It should be emphasized, however, that in these previous studies the residual affinities of ternary gp120–CD4-coreceptor complexes for chemokines were not quantitatively measured. Therefore, it remained theoretically possible that these ternary complexes might prevent down-modulation and be supersensitive rather than inhibited in their abilities to signal in response to chemokines. Our evidence suggests that the net effect of gp120 adsorption is to reduce rather than to amplify signaling in response to chemokines.
Recently, Weissman et al. (16) reported that oligomeric or monomeric gp120s from some M-tropic isolates of HIV-1 can induce CCR5-mediated Ca2+ mobilization in activated B10 lymphocytic cells and in activated CD4-positive T lymphocytes. Notably, however, gp120s from some viruses that use CCR5 as a coreceptor failed to activate signaling, and gp120s from T-cell-tropic HIV-1 isolates did not activate CXCR4. Moreover, only a fraction (ca. 15–30%) of the cells in their assay samples responded to gp120, and these responses generally appeared substantially weaker than responses of the same cells to MIP-1β. In addition, Davis et al. (17) reported that gp120 preparations from T-cell-tropic and M-tropic isolates of HIV-1 increased the phosphorylation of Pyk2 tyrosine kinase but did not induce detectable Ca2+ mobilization in activated CD4-positive T lymphocytes. In contrast, SDF-1α and MIP-1β induced substantial Ca2+ mobilization in the same cells (17). These effects of gp120 are difficult to reconcile because activation of phospholipase C with resultant Ca2+ release is a proximal effect of activated Gβγ-subunits (31, 39), whereas Pyk2 phosphorylation is a downstream response that depends on an increase in cytosolic Ca2+ concentration (21, 22). Consequently, it is likely that gp120s used by Davis et al. (17) induced Ca2+ mobilization to a small but undetectable extent in the cell population, implying that they were only weak CXCR4 and CCR5 agonists or that they functioned as strong agonists in only a small proportion of the chemokine-responsive cells. Together, these results suggest that oligomeric or monomeric gp120s might function to variable extents as weak coreceptor agonists, perhaps only in proliferating cells at one stage of the cell cycle or in the presence of Lck tyrosine kinase or other lymphocyte accessory proteins. Our results are compatible with these interpretations. It is important to emphasize that potential antagonist effects of gp120 were neither examined nor excluded by these previous studies. Conceivably, gp120s could be both weak agonists and strong antagonists of coreceptor signaling, perhaps depending on specific intracellular or extracellular factors such as proteoglycans. Presumably, the structural organization of the virion envelope could also influence the agonist and antagonist activities.
We believe that antagonist effects of oligomeric or monomeric gp120s on CXCR4 and CCR5 responses to chemokines would very likely also occur in the natural cellular targets for HIV-1 infections. Because these antagonisms appear to involve direct interactions of gp120–CD4 complexes with CXCR4 and CCR5 in a common membrane, they would be expected to occur ubiquitously. Accordingly, previous evidence has demonstrated gp120–CD4-coreceptor ternary complexes (10) and gp120- and CD4-mediated competitive displacement of 125I-labeled chemokines from CXCR4 and CCR5 in human lymphocytes and other cells (8, 9, 12, 13). However, because of complexities in the signaling and desensitization pathways of T lymphocytes as described above, and the relatively low rate of gp120 binding onto cell surface CD4 (see Fig. 3), we anticipate that antagonist effects of gp120s in lymphocytes may prove difficult to unambiguously distinguish from weak agonist-induced desensitizations. By either mechanism, however, a major expected consequence of gp120 adsoprtion would be attenuation of responses to specific chemokines.
Based on these considerations, we conclude that gp120s shed from virions and from infected cells may bind to uninfected CD4-positive cells and antagonize their responses to specific chemokines. This could possibly have a major influence on immune and inflammatory responses of infected individuals. Further investigations will be needed to understand the importance of this inhibition and of the recently reported gp120-mediated CXCR4 and CCR5 activations (16, 17) on the development of HIV-1-induced disease.
Acknowledgments
We are grateful to Drs. Shiu-lok Hu, James Arthos, Anthony Fauci, and Raymond Sweet for generous gifts of gp120s. Special thanks to Jacques Wadiche for discussion, Ali Nouri for expert technical assistance, and our colleagues in both laboratories for advice and encouragement. This research was supported by National Institutes of Health Grants CA67358 and NS33270. N.M. is partially supported by a National Institute of Health predoctoral fellowship in Molecular Hematology and Oncology (T32HL07781).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HIV-1, HIV type 1; M-tropic, macrophage-tropic.
References
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 3.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 4.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 5.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 7.D’Souza M P, Harden V A. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 8.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 9.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 11.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 12.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, et al. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 14.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C L, Goldsmith M A, Charo I F. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 16.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Nature (London) 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 17.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folgueira L, Algeciras A, MacMorran W S, Bren G D, Paya C V. J Virol. 1996;70:2332–2338. doi: 10.1128/jvi.70.4.2332-2338.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popik W, Pitha P M. Mol Cell Biol. 1996;16:6532–6541. doi: 10.1128/mcb.16.11.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briand G, Barbeau B, Tremblay M. Virology. 1997;228:171–179. doi: 10.1006/viro.1996.8399. [DOI] [PubMed] [Google Scholar]
- 21.Lev S, Moreno H, Martinez R, Canoll P, Peles E, Musacchio J M, Plowman G D, Rudy B, Schlessinger J. Nature (London) 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- 22.Della Rocca G J, van Biesen T, Daaka Y, Luttrell D K, Luttrell L M, Lefkowitz R J. J Biol Chem. 1997;272:19125–19132. doi: 10.1074/jbc.272.31.19125. [DOI] [PubMed] [Google Scholar]
- 23.Anand-Apte B, Zetter B R, Viswanathan A, Qiu R G, Chen J, Ruggieri R, Symons M. J Biol Chem. 1997;272:30688–30692. doi: 10.1074/jbc.272.49.30688. [DOI] [PubMed] [Google Scholar]
- 24.Dikic I, Tokiwa G, Lev S, Courtneidge S A, Schlessinger J. Nature (London) 1996;383:547–550. doi: 10.1038/383547a0. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Ilasaca M, Crespo P, Pellici P G, Gutkind J S, Wetzker R. Science. 1997;275:394–397. doi: 10.1126/science.275.5298.394. [DOI] [PubMed] [Google Scholar]
- 26.Kubo Y, Reuveny E, Slesinger P A, Jan Y N, Jan L Y. Nature (London) 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 27.Dascal N, Schreibmayer W, Lim N F, Wang W, Chavkin C, DiMagno L, Labarca C, Kieffer B L, Gaveriaux-Ruff C, Trollinger D, et al. Proc Natl Acad Sci USA. 1993;90:10235–10239. doi: 10.1073/pnas.90.21.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim N F, Dascal N, Labarca C, Davidson N, Lester H A. J Gen Physiol. 1995;105:421–439. doi: 10.1085/jgp.105.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuveny E, Slesinger P A, Inglese J, Morales J M, Iniguez-Lluhi J A, Lefkowitz R J, Bourne H R, Jan Y N, Jan L Y. Nature (London) 1994;370:143–146. doi: 10.1038/370143a0. [DOI] [PubMed] [Google Scholar]
- 30.Schreibmayer W, Dessauer C W, Vorobiov D, Gilman A G, Lester H A, Davidson N, Dascal N. Nature (London) 1996;380:624–627. doi: 10.1038/380624a0. [DOI] [PubMed] [Google Scholar]
- 31.Clapham D E. Annu Rev Neurosci. 1994;17:441–464. doi: 10.1146/annurev.ne.17.030194.002301. [DOI] [PubMed] [Google Scholar]
- 32.Kovoor A, Henry D J, Chavkin C. J Biol Chem. 1995;270:589–595. doi: 10.1074/jbc.270.2.589. [DOI] [PubMed] [Google Scholar]
- 33.Doupnik C A, Lim N F, Kofuji P, Davidson N, Lester H A. J Gen Physiol. 1995;106:1–23. doi: 10.1085/jgp.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saugstad J A, Segerson T P, Westbrook G L. J Neurosci. 1996;16:5979–5985. doi: 10.1523/JNEUROSCI.16-19-05979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabat D, Kozak S L, Wehrly K, Chesebro B. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhmann S E, Platt E J, Kozak S L, Kabat D. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacon K B, Premack B A, Gardner P, Schall T J. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H, Kuang Y, Wu Y, Smrcka A, Simon M I, Wu D. J Biol Chem. 1996;271:13430–13434. doi: 10.1074/jbc.271.23.13430. [DOI] [PubMed] [Google Scholar]
- 40.Prado G N, Suzuki H, Wilkinson N, Cousins B, Navarro J. J Biol Chem. 1996;271:19186–19190. doi: 10.1074/jbc.271.32.19186. [DOI] [PubMed] [Google Scholar]
- 41.Mueller S G, White J R, Schraw W P, Lam V, Richmond A. J Biol Chem. 1997;272:8207–8214. doi: 10.1074/jbc.272.13.8207. [DOI] [PubMed] [Google Scholar]
- 42.Haribabu B, Richardson R M, Fisher I, Sozzani S, Peiper S C, Horuk R, Ali H, Snyderman R. J Biol Chem. 1997;272:28726–28731. doi: 10.1074/jbc.272.45.28726. [DOI] [PubMed] [Google Scholar]
- 43.Solari R, Offord R E, Remy S, Aubry J P, Wells T N, Whitehorn E, Oung T, Proudfoot A E. J Biol Chem. 1997;272:9617–9620. doi: 10.1074/jbc.272.15.9617. [DOI] [PubMed] [Google Scholar]
- 44.Barak L S, Ferguson S S G, Zhang J, Caron M G. J Biol Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]