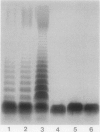
Abstract
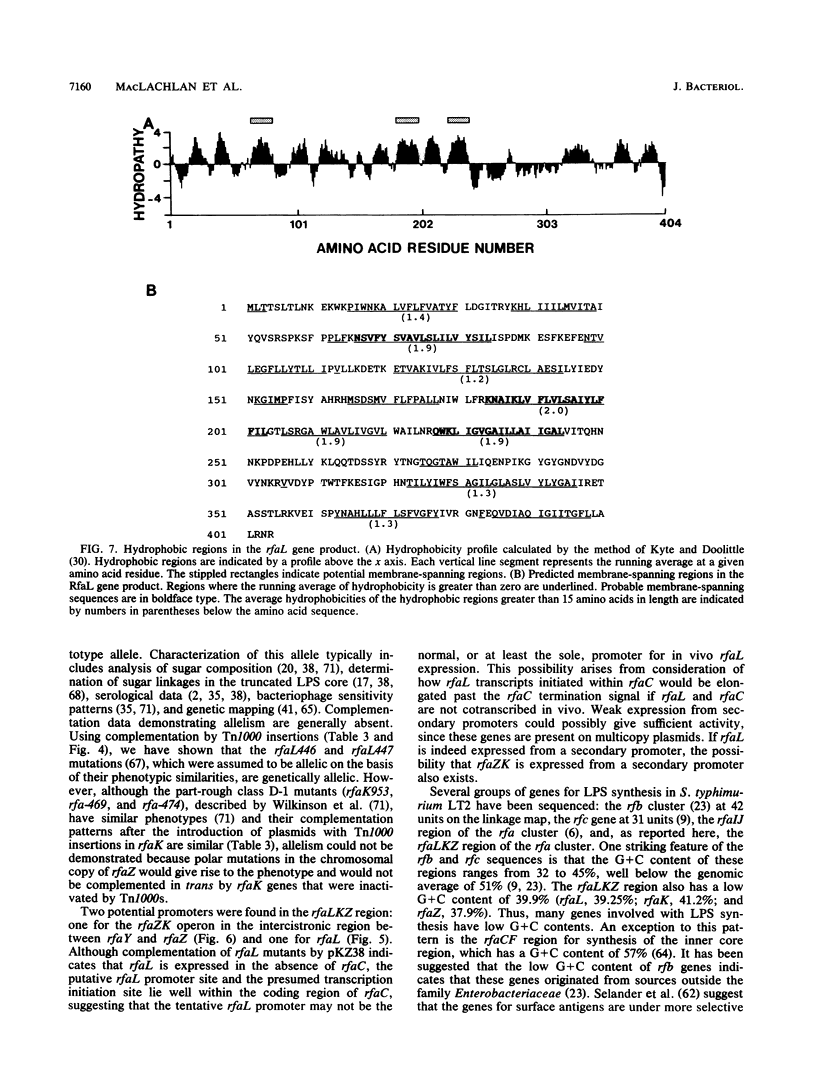
We have cloned and sequenced the rfaL and rfaK genes for lipopolysaccharide synthesis in Salmonella typhimurium LT2 on a 4.28-kb HindIII fragment from the previously described R' factor pKZ3 (S. K. Kadam, A. Rehemtulla, and K. E. Sanderson, J. Bacteriol. 161:277-284, 1985). rfaL is thought to encode a component of the O-antigen ligase, and rfaK is believed to encode the N-acetylglucosamine transferase. The genes were identified by the loss of complementation of prototype rfaL and rfaK mutations after Tn1000 mutagenesis. Translation of the nucleotide sequence predicted sizes of 45.9 and 43.1 kDa for the rfaL and rfaK gene products, respectively. Hydropathy analysis of the rfaL product suggested that it was an integral membrane protein. A third gene, rfaZ, was found to be an 808-bp open reading frame on the pyrE side of rfaK. Insertions into rfaZ reduced rfaK complementation, suggesting cotranscription in the pyrE-cysE direction. The rfaL gene is transcribed in the opposite direction in a separate operon which may also include rfaC. An incomplete open reading frame with homology to an Escherichia coli gene in the same region, rfaY, was found on the pyrE side of rfaZ. Complementation studies with Tn1000 insertions in rfaL showed that rfaL446 and rfaL447 are allelic. With the cloning of the rfaL and -K genes, the order of genes within the rfa cluster at 79 units on the linkage map was found to be cysE-rfaDFCLKZYJIBG-pyrE.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin E. A., Graves J. F., Hite L. A., Parker C. T., Schnaitman C. A. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J Bacteriol. 1990 Sep;172(9):5312–5325. doi: 10.1128/jb.172.9.5312-5325.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECKMANN I., SUBBAIAH T. V., STOCKER B. A. ROUGH MUTANTS OF SALMONELLA TYPHIMURIUM. II. SEROLOGICAL AND CHEMICAL INVESTIGATIONS. Nature. 1964 Mar 28;201:1299–1301. doi: 10.1038/2011299a0. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bullas L. R., Ryu J. I. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983 Oct;156(1):471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstenius P., Flock J. I., Lindberg A. Nucleotide sequence of rfaI and rfaJ genes encoding lipopolysaccharide glycosyl transferases from Salmonella typhimurium. Nucleic Acids Res. 1990 Oct 25;18(20):6128–6128. doi: 10.1093/nar/18.20.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Niemela S. L., Miller R. H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L. V., Hackett J. Molecular cloning, characterization, and nucleotide sequence of the rfc gene, which encodes an O-antigen polymerase of Salmonella typhimurium. J Bacteriol. 1991 Apr;173(8):2521–2529. doi: 10.1128/jb.173.8.2521-2529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Rothfield L. Studies of a phospholipid-requiring bacterial enzyme. I. Purification and properties of uridine diphosphate galactose: lipopolysaccharide alpha-3-galactosyl transferase. Biochemistry. 1969 Sep;8(9):3500–3507. doi: 10.1021/bi00837a003. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. Uses of the transposon gamma delta in the analysis of cloned genes. Methods Enzymol. 1983;101:362–369. doi: 10.1016/0076-6879(83)01027-7. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander I. M., Vaara M., Sukupolvi S., Rhen M., Saarela S., Zähringer U., Mäkelä P. H. rfaP mutants of Salmonella typhimurium. Eur J Biochem. 1989 Nov 20;185(3):541–546. doi: 10.1111/j.1432-1033.1989.tb15147.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Jiang X. M., Neal B., Santiago F., Lee S. J., Romana L. K., Reeves P. R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol Microbiol. 1991 Mar;5(3):695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Jousimies H., Mäkelä P. H. Genetic analysis of Salmonella minnesota R mutants with defects in the biosynthesis of the lipopolysaccharide core. J Bacteriol. 1974 Sep;119(3):753–759. doi: 10.1128/jb.119.3.753-759.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S. K., Peppler M. S., Sanderson K. E. Temperature-sensitive mutants in rfaI and rfaJ, genes for galactosyltransferase I and glucosyltransferase II, for synthesis of lipopolysaccharide in Salmonella typhimurium. Can J Microbiol. 1985 Sep;31(9):861–869. doi: 10.1139/m85-160. [DOI] [PubMed] [Google Scholar]
- Kadam S. K., Rehemtulla A., Sanderson K. E. Cloning of rfaG, B, I, and J genes for glycosyltransferase enzymes for synthesis of the lipopolysaccharide core of Salmonella typhimurium. J Bacteriol. 1985 Jan;161(1):277–284. doi: 10.1128/jb.161.1.277-284.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kleffel B., Garavito R. M., Baumeister W., Rosenbusch J. P. Secondary structure of a channel-forming protein: porin from E. coli outer membranes. EMBO J. 1985 Jun;4(6):1589–1592. doi: 10.1002/j.1460-2075.1985.tb03821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. T., Stocker B. A. Mapping of rfa Genes in Salmonella typhimurium by ES18 and P22 Transduction and by Conjugation. J Bacteriol. 1972 Oct;112(1):48–57. doi: 10.1128/jb.112.1.48-57.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laskowski M., Sr Purification and properties of the mung bean nuclease. Methods Enzymol. 1980;65(1):263–276. doi: 10.1016/s0076-6879(80)65036-8. [DOI] [PubMed] [Google Scholar]
- Lehmann V., Hämmerling G., Nurminen M., Minner I., Ruschmann E., Lüderitz O., Kuo T. T., Stocker B. A. A new class of heptose-defective mutant of Salmonella typhimurium. Eur J Biochem. 1973 Jan 15;32(2):268–275. doi: 10.1111/j.1432-1033.1973.tb02607.x. [DOI] [PubMed] [Google Scholar]
- Lesse A. J., Campagnari A. A., Bittner W. E., Apicella M. A. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods. 1990 Jan 24;126(1):109–117. doi: 10.1016/0022-1759(90)90018-q. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Hellerqvist C. G. Bacteriophage attachment sites, serological specificity, and chemical composition of the lipopolysaccharides of semirough and rough mutants of Salmonella typhimurium. J Bacteriol. 1971 Jan;105(1):57–64. doi: 10.1128/jb.105.1.57-64.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. A., Hellerqvist C. G. Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J Gen Microbiol. 1980 Jan;116(1):25–32. doi: 10.1099/00221287-116-1-25. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Holme T. Influence of O side chains on the attachment of the Felix O-1 bacteriophage to Salmonella bacteria. J Bacteriol. 1969 Aug;99(2):513–519. doi: 10.1128/jb.99.2.513-519.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan P. R., Sanderson K. E. Transformation of Salmonella typhimurium with plasmid DNA: differences between rough and smooth strains. J Bacteriol. 1985 Jan;161(1):442–445. doi: 10.1128/jb.161.1.442-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath B. C., Osborn M. J. Localization of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J Bacteriol. 1991 Jan;173(2):649–654. doi: 10.1128/jb.173.2.649-654.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller E., Hinckley A., Rothfield L. Studies of phospholipid-requiring bacterial enzymes. 3. Purification and properties of uridine diphosphate glucose:lipopolysaccharide glucosyltransferase I. J Biol Chem. 1972 Apr 25;247(8):2614–2622. [PubMed] [Google Scholar]
- Osborn M. J. Biochemical characterization of mutants of Salmonella typhimurium lacking glucosyl or galactosyl lipopolysaccharide transferases. Nature. 1968 Mar 9;217(5132):957–960. doi: 10.1038/217957a0. [DOI] [PubMed] [Google Scholar]
- Osborn M. J., D'Ari L. Enzymatic incorporation of N-acetylglucosamine into cell wall lipopolysaccharide in a mutant strain of Salmonella typhimurium. Biochem Biophys Res Commun. 1964 Aug 11;16(6):568–575. doi: 10.1016/0006-291x(64)90194-9. [DOI] [PubMed] [Google Scholar]
- Osborn M. J., Gander J. E., Parisi E. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. J Biol Chem. 1972 Jun 25;247(12):3973–3986. [PubMed] [Google Scholar]
- Portnoy D. A., Moseley S. L., Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981 Feb;31(2):775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C. R. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roantree R. J., Kuo T. T., MacPhee D. G. The effect of defined lipopolysaccharide core defects upon antibiotic resistances of Salmonella typhimurium. J Gen Microbiol. 1977 Dec;103(2):223–234. doi: 10.1099/00221287-103-2-223. [DOI] [PubMed] [Google Scholar]
- SUBBAIAH T. V., STOCKER B. A. ROUGH MUTANTS OF SALMONELLA TYPHIMURIUM. I. GENETICS. Nature. 1964 Mar 28;201:1298–1299. doi: 10.1038/2011298a0. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Stocker B. A. Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J Bacteriol. 1981 May;146(2):535–541. doi: 10.1128/jb.146.2.535-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Van Wyngaarden J., Lüderitz O., Stocker B. A. Rough mutants of Salmonella typhimurium with defects in the heptose region of the lipopolysaccharide core. Can J Microbiol. 1974 Aug;20(8):1127–1134. doi: 10.1139/m74-175. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. W., Lüderitz O., Westphal O. Studies on the structure of lipopolysaccharides of Salmonella minnesota and Salmonella typhimurium R strains. Biochem J. 1965 Aug;96(2):439–448. doi: 10.1042/bj0960439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Wilkinson R. G., Gemski P., Jr, Stocker B. A. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972 May;70(3):527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- Wollin R., Creeger E. S., Rothfield L. I., Stocker B. A., Lindberg A. A. Salmonella typhimurium mutants defective in UDP-D-galactose:lipopolysaccharide alpha 1,6-D-galactosyltransferase. Structural, immunochemical, and enzymologic studies of rfaB mutants. J Biol Chem. 1983 Mar 25;258(6):3769–3774. [PubMed] [Google Scholar]
- YAMAMOTO N., ANDERSON T. F. Genomic masking and recombination between serologically unrelated phages P22 and P221. Virology. 1961 Aug;14:430–439. doi: 10.1016/0042-6822(61)90334-8. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- de Lencastre H., Chak K. F., Piggot P. J. Use of the Escherichia coli transposon Tn1000 (gamma delta) to generate mutations in Bacillus subtilis DNA. J Gen Microbiol. 1983 Oct;129(10):3203–3210. doi: 10.1099/00221287-129-10-3203. [DOI] [PubMed] [Google Scholar]