Abstract
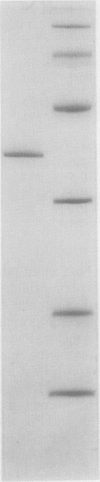
Hydroxypyruvate reductase was purified to homogeneity from the facultative methylotroph Methylobacterium extorquens AM1. It has a molecular mass of about 71 kDa, and it consists of two identical subunits with a molecular mass of about 37 kDa. This enzyme uses both NADH (Km = 0.04 mM) and NADPH (Km = 0.06 mM) as cofactors, uses hydroxypyruvate (Km = 0.1 mM) and glyoxylate (Km = 1.5 mM) as the only substrates for the forward reaction, and carries out the reverse reaction with glycerate (Km = 2.6 mM) only. It was not possible to detect the conversion of glycolate to glyoxylate, a proposed role for this enzyme. Kinetics and inhibitory studies of the enzyme from M. extorquens AM1 suggest that hydroxypyruvate reductase is not a site for regulation of the serine cycle at the level of enzyme activity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Lidstrom M. E. The moxFG region encodes four polypeptides in the methanol-oxidizing bacterium Methylobacterium sp. strain AM1. J Bacteriol. 1988 May;170(5):2254–2262. doi: 10.1128/jb.170.5.2254-2262.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The role of glyoxylate, glycollate and acetate in the growth of Pseudomonas AM1 on ethanol and on C 1 compounds. Biochem J. 1972 Jun;128(1):107–115. doi: 10.1042/bj1280107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husic D. W., Tolbert N. E. NADH:hydroxypyruvate reductase and NADPH:glyoxylate reductase in algae: partial purification and characterization from Chlamydomonas reinhardtii. Arch Biochem Biophys. 1987 Feb 1;252(2):396–408. doi: 10.1016/0003-9861(87)90046-4. [DOI] [PubMed] [Google Scholar]
- Izumi Y., Yoshida T., Kanzaki H., Toki S., Miyazaki S. S., Yamada H. Purification and characterization of hydroxypyruvate reductase from a serine-producing methylotroph, Hyphomicrobium methylovorum GM2. Eur J Biochem. 1990 Jun 20;190(2):279–284. doi: 10.1111/j.1432-1033.1990.tb15573.x. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Kohn L. D., Jakoby W. B. Tartaric acid metabolism. VII. Crystalline hydroxypyruvate reductase (D-glycerate dehydrogenase). J Biol Chem. 1968 May 25;243(10):2494–2499. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Large P. J., Quayle J. R. Microbial growth on C(1) compounds. 5. Enzyme activities in extracts of Pseudomonas AM1. Biochem J. 1963 May;87(2):386–396. doi: 10.1042/bj0870386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom M. E., Stirling D. I. Methylotrophs: genetics and commercial applications. Annu Rev Microbiol. 1990;44:27–58. doi: 10.1146/annurev.mi.44.100190.000331. [DOI] [PubMed] [Google Scholar]
- McNerney T., O'connor M. L. Regulation of enzymes associated with C-1 metabolism in three facultative methylotrophs. Appl Environ Microbiol. 1980 Aug;40(2):370–375. doi: 10.1128/aem.40.2.370-375.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):591–597. doi: 10.1128/jb.166.2.591-597.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I. J., Anthony C. Acetyl-CoA production and utilization during growth of the facultative methylotroph Pseudomonas AM1 on ethanol, malonate and 3-hydroxybutyrate. J Gen Microbiol. 1976 Jul;95(1):134–143. doi: 10.1099/00221287-95-1-134. [DOI] [PubMed] [Google Scholar]
- Titus D. E., Hondred D., Becker W. M. Purification and characterization of hydroxypyruvate reductase from cucumber cotyledons. Plant Physiol. 1983 Jun;72(2):402–408. doi: 10.1104/pp.72.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N. E., Yamazaki R. K., Oeser A. Localization and properties of hydroxypyruvate and glyoxylate reductases in spinach leaf particles. J Biol Chem. 1970 Oct 10;245(19):5129–5136. [PubMed] [Google Scholar]
- Utting J. M., Kohn L. D. Structural, kinetic, and renaturation properties of an induced hydroxypyruvate reductase from Pseudomonas acidovorans. J Biol Chem. 1975 Jul 10;250(13):5233–5242. [PubMed] [Google Scholar]
- WILLIS J. E., SALLACH H. J. Evidence for a mammalian D-glyceric dehydrogenase. J Biol Chem. 1962 Mar;237:910–915. [PubMed] [Google Scholar]
- Whitaker J. R., Granum P. E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980 Nov 15;109(1):156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]