Abstract
We have analyzed crystal structures of cytochrome bc1 complexes with electron transfer inhibitors bound to the ubiquinone binding pockets Qi and/or Qo in the cytochrome b subunit. The presence or absence of the Qi inhibitor antimycin A did not affect the binding of the Qo inhibitors. Different subtypes of Qo inhibitors had dramatically different effects on the mobility of the extramembrane domain of the iron–sulfur protein (ISP): Binding of 5-undecyl-6-hydroxy-4,7-dioxobenzothiazol and stigmatellin (subtype Qo–II and Qo–III, respectively) led to a fixation of the ISP domain on the surface of cytochrome b, whereas binding of myxothiazol and methoxyacrylate-stilbene (subtype Qo–I) favored release of this domain. The native structure has an empty Qo pocket and is intermediate between these extremes. On the basis of these observations we propose a model of quinone oxidation in the bc1 complex, which incorporates fixed and loose states of the ISP as features important for electron transfer and, possibly, also proton transport.
Ubiquinol–cytochrome-c oxidoreductase (cytochrome bc1 complex; EC 1.10.2.2) is a segment of the respiratory chain in mitochondria and of the photosynthetic apparatus of purple bacteria. It catalyzes electron transfer (ET) from ubiquinol to cytochrome c, coupled to proton transport across a membrane (from the matrix space to the intermembrane space of mitochondria; from the cytoplasm to the periplasm of purple bacteria). The resulting electrochemical proton gradient drives ATP synthesis and transport processes (1, 2). Essential for the function of the bc1 complex are the three redox proteins cytochrome b, cytochrome c1, and the iron–sulfur protein (ISP). Two b-type hemes (bL and bH) are attached to cytochrome b, one c-type heme is bound to cytochrome c1, and a Rieske-type iron–sulfur center (FeS) is bound to ISP (2). Whereas some bacterial bc1 complexes consist of only those redox subunits (3), their mitochondrial counterparts contain up to 8 additional protein subunits whose precise functions in the complex are largely unknown (4).
The protonmotive Q cycle model (2, 5, 6) best explains experimental results on the ET pathway through the four redox centers of the bc1 complex. It postulates two separate ubiquinone binding sites, called Qo and Qi. In bc1 complexes of the inner membrane of mitochondria, Qo is located near the membrane surface facing the intermembrane space, and Qi is near the membrane surface facing the matrix space. The Q cycle model requires bifurcated electron flow from ubiquinol bound in the Qo site: The first electron of ubiquinol is sequentially transferred to the ISP, cytochrome c1, and eventually to the soluble cytochrome c. Protons are released into the intermembrane space, generating a ubisemiquinone anion in the Qo site. The second electron is transferred to hemes bL and bH and to a ubiquinone or a ubisemiquinone anion in the Qi site. The fully reduced quinone in the Qi site picks up two protons from the matrix space and moves to the Qo site for recycling.
The discovery of different types of specific ET inhibitors of the bc1 complex was crucial for the development of this Q cycle hypothesis. The two major types of bc1 inhibitors are called Qo or Qi inhibitors, depending on their action in the cytochrome bc1 complex (7, 8). All Qi inhibitors target specifically the ET path from heme bL to ubiquinone/ubisemiquinone in the Qi site; they do not share common structural motifs. Qo inhibitors block binding of quinol to the Qo site and ET through this site. They can be classified further on the basis of common structural motifs and of their effects on the absorption spectrum of heme bL and on the electron paramagnetic resonance (EPR) spectrum and redox potential of the FeS (7). One Qo inhibitor subtype shares a methoxyacrylate (MOA) group (examples: myxothiazol, MOA-stilbene), another subtype resembles a hydroxyquinone molecule (example: 5-undecyl-6-hydroxy-4,7-dioxobenzothiazol or UHDBT), and a third subtype has a chromone group as the common motif (example: stigmatellin). MOA inhibitors cause solely a red-shift of the bL heme’s spectrum (9) and hydroxyquinone inhibitors alter the EPR spectrum of the FeS (10), whereas the chromone inhibitors affect both spectra (11).
The possibility of two inhibitor binding sites in the Qo pocket has led to the so-called “catalytic switch” model (12). This model assumed at least two conformational states of the Qo pocket in cytochrome b, which are directly controlled by the redox state of the ISP. By alternating between these two conformational states, bifurcated ET is enforced during the catalytic reaction. Separately, the “double Qo occupancy” model was recently proposed; it is based on the observation of two distinct EPR signals that depend on the redox state or on ubiquinone concentration in bacterial bc1 complexes. According to this model, the Qo pocket has two ubiquinone binding sites with different affinities (13, 14). Later, this became one of the foundations of the “proton-gated charge-transfer” model that integrates ET and proton translocation (15).
Recently, the crystal structure of the bovine bc1 complex was reported, and the binding sites of the Qi inhibitor antimycin A and of the Qo inhibitor myxothiazol were identified (16). The two binding sites are formed by the cytochrome b subunit near opposite membrane surfaces. They are close to iron positions determined from anomalous scattering data. Here, we report further crystallographic studies of inhibitor binding to the bc1 complex and studies of the effects of different types of inhibitors on the ISP and the location of its FeS.
MATERIALS AND METHODS
Protein Preparation.
Cytochrome bc1 complex from bovine heart mitochondria was prepared, starting from highly purified succinate–cytochrome-c reductase (17) as described previously (16, 18). bc1 particles were solubilized by deoxycholate, and contaminants were removed by a 15-step ammonium acetate fractionation. Pure bc1 complex in the oxidized form was recovered from the precipitates formed between 18.5% and 33.5% saturation with ammonium acetate. For crystallization, this material was adjusted to a final concentration of 20 mg/ml in a solution containing 50 mM Mops buffer at pH 7.2, 20 mM ammonium acetate, 20% (wt/vol) glycerol, and 0.1% either decanoyl-N-methylglucamide or diheptanoyl phosphatidylcholine.
Cocrystallization with Inhibitors.
For cocrystallization of the bc1 complex with various inhibitors, a 2-fold molar excess of inhibitors was added to the protein solution. This solution was set up for crystallization as described (16, 18). Crystals grew in 3–4 weeks; they had a rectangular shape and ranged in size from 0.4 mm to 0.7 mm. They could be frozen at high glycerol concentration, and they had the same symmetry (space group I4122) and similar unit cell dimensions as the native crystals: a = b = 153.5 Å, c = 597.7 Å.
Data Collection and Analysis.
When flash frozen and kept at 100 K, the crystals were stable enough in strong x-ray beams to allow collection of complete sets of diffraction data. Data were collected on imaging plates at beamlines X4A, X12B, and X25 of the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory, beamline BL4 of the European Synchrotron Radiation Facility (ESRF), and at beamline 7-1 of Stanford Synchrotron Radiation Laboratory (SSRL). The raw diffraction data were processed with the denzo/hkl package (19); Bijvoet pairs were kept separated. Programs mtzutils, scaleit, and fft from the CCP4 package (20) were used for merging and scaling the data with the native data and for the calculation of difference-Fourier maps. Phases for the structure factors of inhibitor-bound crystals in the resolution range 20–3.0 Å were calculated, starting from the native multiple isomorphous replacement phase set (20- to 3.5-Å resolution), by density modification and phase extension in small steps [program dm (21)]. The position and orientation of the extramembrane domain of the ISP were determined by searching electron density maps (20- to 3.0-Å resolution), using the high-resolution structure of this domain (22) as a search model. The search procedure (D.X. et al., unpublished work) calculated linear correlation coefficients between map density and model density under variation of the model orientation or position. First, the FeS of the model was translated to the experimentally determined cluster position in the crystal and the model was systematically rotated around this position. The orientation of maximal correlation was further refined by alternating scans of small shift and orientation ranges until convergence was reached.
RESULTS
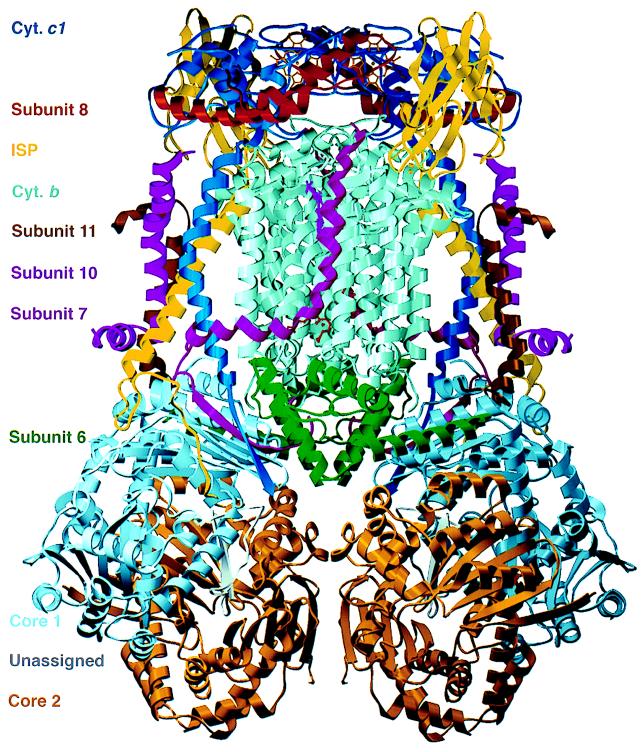
Fig. 1 shows an overall view of our current model of the dimeric bc1 complex. Crystallographic refinement of this model is in progress and will be reported elsewhere; the model includes 15,600 non-hydrogen atoms; the values of Rwork and Rfree in the resolution range of 20–2.7 Å are 0.313 and 0.378, respectively. Phases calculated from this model without the ISP extramembrane domain were used in our real-space searches for the orientation of this domain.
Figure 1.
Ribbon model of the dimeric cytochrome bc1 complex. Colors identifying the subunits are given at the left margin. An as-yet-unassigned peptide is bound in a cavity formed by subunits core 1 and core 2 (16). Figs. 1 and 5 were prepared with the programs molscript (33) and setor (34), respectively.
We cocrystallized the bovine bc1 complex with the Qi inhibitor antimycin A and with the Qo inhibitors myxothiazol, MOA-stilbene, UHDBT, and stigmatellin, and with combinations of antimycin A and each of the Qo inhibitors. The crystallization behavior of the bc1 complex mixed with inhibitors was similar to that of the native protein (18); the cocrystals were isomorphous with the native crystals, and the x-ray diffraction data obtained from these crystals were of comparable quality. Table 1 lists statistics up to 3.0-Å resolution of the diffraction data sets used in this study; some of the data sets, including Native 1, extend well beyond that resolution. Using multiple isomorphous replacement phases improved by solvent flattening (16), we analyzed each cocrystal by calculating difference-density maps (structure factor amplitudes: |FI| − |FN|, where |FI| and |FN| are amplitudes of inhibitor-containing crystals and native crystals, respectively) and anomalous difference maps (structure factor amplitudes: |F+| − |F−|, where |F+| and |F−| are amplitudes of Bijvoet pairs of reflections from the same crystal). In all cases, the difference-density maps clearly showed electron density for the inhibitors (Fig. 2). Binding of the Qo inhibitors was independent of the presence or absence of the Qi inhibitor, and vice versa. This result was expected because of the spatial separation of the binding sites near opposite membrane surfaces (16), and it is consistent with the experimental observation of additive inhibitory activity of these two major inhibitor types.
Table 1.
X-ray diffraction data statistics (dmin ≥ 3.0 Å)
| Data set | λ, Å | dmin, Å | Unique reflections | Observations | Mean redundancy | Completeness, % | Mosaicity, ° | I/σ(I) | Rmerge, % |
|---|---|---|---|---|---|---|---|---|---|
| Native 1* | 1.01 | 3.0 | 65,901 | 1,715,432 | 26.0 | 91.6 | 0.620 | 16.2 | 14.0 |
| Native 2† | 1.74 | 3.2 | 43,969 | 601,180 | 13.7 | 74.7 | 0.555 | 12.7 | 9.8 |
| Antimycin A‡ | 1.55 | 3.4 | 45,286 | 586,286 | 12.9 | 89.9 | 0.602 | 14.9 | 10.7 |
| MOA-stilbene§ | 0.99 | 3.0 | 63,789 | 1,054,638 | 16.5 | 89.9 | 0.750 | 19.0 | 4.9 |
| UHDBT¶ | 1.08 | 3.0 | 68,766 | 1,382,177 | 20.1 | 96.8 | 0.727 | 12.0 | 11.2 |
| Stigmatellin§ | 0.99 | 3.0 | 50,387 | 736,039 | 14.6 | 68.8 | 0.678 | 6.2 | 19.6 |
| Antimycin A + MOA-stilbene† | 1.74 | 3.3 | 56,192 | 809,082 | 14.4 | 92.5 | 0.394 | 15.4 | 11.0 |
| Antimycin A + myxothiazol† | 1.74 | 3.0 | 65,729 | 940,740 | 14.3 | 92.2 | 0.474 | 7.6 | 14.7 |
| Antimycin A + UHDBT† | 1.74 | 3.5 | 45,755 | 808,324 | 17.7 | 96.6 | 0.628 | 11.8 | 13.1 |
| Antimycin A + stigmatellin¶ | 1.08 | 3.3 | 51,414 | 554,575 | 10.8 | 94.8 | 0.564 | 16.0 | 7.4 |
X-ray data were collected from the beamlines X25(∗), X12B(‡), and X4A(†) of the Brookhaven National Laboratory, BL4 (§) of the European Synchrotron Radiation Facility, and 7-1(¶) of the Stanford Synchrotron Radiation Laboratory.
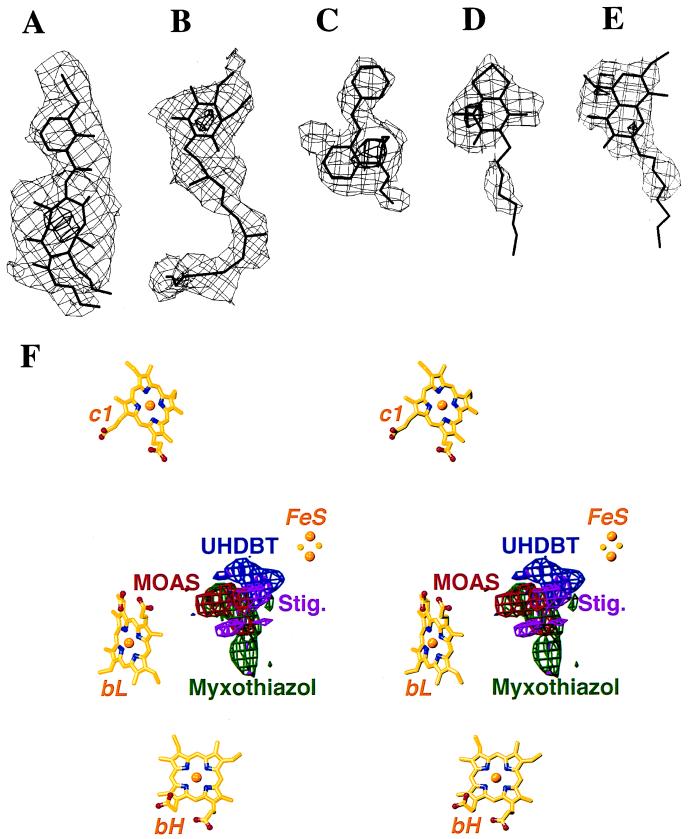
Figure 2.
(A–E) Difference electron densities between the inhibitor-bound and native crystals and atomic models of inhibitors and ubiquinone. The maps are contoured at two levels (thin and thick lines, respectively); contour levels for each map are listed in standard deviations from the mean. (A) Antimycin A (3, 20). (B) Ubiquinone (−3, −10). (C) MOA-stilbene (3, 8). (D) UHDBT (3, 7). (E) Stigmatellin (3, 5). (F) Stereo drawing: Models of redox cofactors together with superimposed difference-densities of four Qo inhibitors, viewed parallel to the membrane. MOAS, MOA-stilbene; Stig., stigmatellin.
Antimycin A.
The difference-density maps contained strong positive density for antimycin A itself near heme bH (Figs. 2A and 5). Except for local changes near the antimycin A binding site, the maps did not indicate any antimycin A-induced change elsewhere in the bc1 structure. In particular, the presence of antimycin A did not influence the positions and relative heights of the peaks representing the iron centers in the anomalous difference maps (Table 2). Strong negative density appeared next to the positive density for antimycin A; its elongated shape resembled that of a ubiquinone molecule (Fig. 2B). Difference-density maps between native crystals soaked with either ubiquinone-10 or ubiquinone-6, and native crystal, had their highest peaks at this site (data not shown). We assume that a ubiquinone molecule was bound in the native structure but displaced by antimycin A; this assumption is consistent with spectroscopic observations (23). It appears that the Qi binding site is not identical to the antimycin A binding site, but close enough for overlap, with antimycin A reaching deeper into the binding pocket.
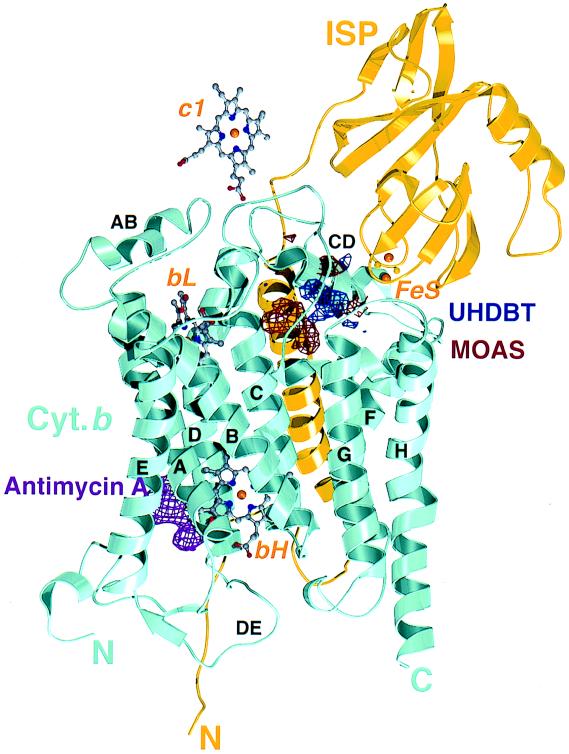
Figure 5.
Cytochrome b and ISP, hemes, and difference densities for MOA-stilbene (MOAS), UHDBT, and antimycin A, viewed parallel to the membrane. The eight transmembrane helices of cytochrome b are labeled A to H; some of the connecting loops are labeled too. The loop CD consists of two antiparallel helices. The structure of the extramembrane domain of ISP is based on the crystal structure of this domain (22), positioned and oriented by using UHDBT data (Tables 1–3); the transmembrane helix of ISP contacts cytochrome b of the second monomer in the dimer (not shown).
Table 2.
Peaks in anomalous difference maps
| Data set | Maximum peak height, units of σ | Normalized peak height*
|
||||
|---|---|---|---|---|---|---|
| bH | bL | c1 | FeS† | Next-highest peak | ||
| Native 1 | 14.7 | 1.00 | 0.99 | 0.53 | 0.45 | 0.28 |
| Native 2 | 27.6 | 1.00 | 0.93 | 0.61 | 0.47 | 0.18 |
| Antimycin A | 14.3 | 1.00 | 1.09 | 0.78 | 0.60 | 0.40 |
| MOA-stilbene | 14.2 | 1.00 | 1.02 | 0.76 | — | 0.53‡ |
| UHDBT | 13.3 | 1.00 | 0.89 | 0.61 | 0.97 | 0.34 |
| Stigmatellin | 18.8 | 1.00 | 0.99 | 0.79 | 1.18 | 0.28 |
| Antimycin A + MOA-stilbene | 29.2 | 1.00 | 0.96 | 0.57 | — | 0.20‡ |
| Antimycin A + myxothiazol | 20.1 | 1.00 | 0.97 | 0.66 | 0.38 | 0.24 |
| Antimycin A + UHDBT | 21.8 | 1.00 | 0.77 | 0.60 | 0.83 | 0.32 |
| Antimycin A + stigmatellin | 13.3 | 1.00 | 0.95 | 0.56 | 1.25 | 0.36 |
Normalization to bH peak.
FeS position in the fixed state.
Possible alternative position of FeS in the loose state.
Qo Site Inhibitors.
The Qo inhibitors occupy different subsites in the Qo pocket. Except for the combination MOA-stilbene/UHDBT, their binding sites overlap, which explains why binding of these Qo inhibitors is mutually exclusive (24, 25). Myxothiazol and MOA-stilbene bind close to the heme bL, UHDBT binds close to the FeS, and stigmatellin overlaps both binding sites (Figs. 2F and 5). This finding is in perfect agreement with the aforementioned spectroscopic changes caused by binding of these inhibitors to the bc1 complex. Different binding regions for different Qo inhibitors were also predicted from functional studies on inhibitor-resistant mutants (26–29). On the basis of these observations, the Qo inhibitors used in this study can be divided into three categories: Qo-I (myxothiazol, MOA-stilbene), Qo-II (UHDBT), and Qo-III (stigmatellin) (7).
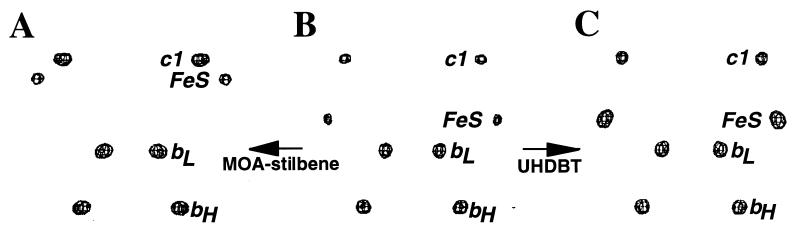
Binding of Qo inhibitors had considerable influence on the heights and positions of peaks corresponding to the FeS in the anomalous difference maps. More importantly, there was a strong correlation between the change in this anomalous signal (compared with that of native crystals) and the subtype of the Qo inhibitor. Binding of MOA-stilbene (type Qo-I) abolished the anomalous signal for the FeS at the position observed in the native crystals (Table 2). Instead, a minor peak appeared closer to cytochrome c1, 15 Å away from the native position (Fig. 3A). Although small compared with the other peaks, it appeared consistently in all forms of MOA-stilbene-containing crystals (Table 2), and it may represent a weakly occupied alternative position of the FeS. Most likely, the changes in the FeS signal indicate increased mobility of the extramembrane domain of the ISP (22), which, because of its rigidity, would be able to perform a hinged motion relative to its transmembrane domain. Positive and negative difference-density between native crystals and the inhibitor cocrystals, spread over the entire ISP region, indicated a conformational change of the whole extramembrane domain of the ISP (data not shown). The absence of difference-density in the transmembrane region of the ISP showed that the ISP subunit was not accidentally lost from the complex.
Figure 3.
Anomalous difference maps at 5-Å resolution of crystals containing MOA-stilbene (A), no inhibitor (native) (B), and UHDBT (C); the four redox centers are labeled. The maps are contoured at five standard deviations above the mean. The peak for the FeS in the UHDBT is stronger than the one in the native structure but becomes equivalent in the UHDBT-bound structure (Table 2).
Myxothiazol, which belongs to the same Qo-I subtype as MOA-stilbene, causes a significant lowering, but not the disappearance, of the anomalous signal for the FeS (Table 2). If the strength of this signal is a measure of the mobility of the ISP extramembrane domain, the effects of MOA-stilbene and myxothiazol on this mobility go in the same direction, with MOA-stilbene being the more potent of the two.
In contrast to the Qo-I inhibitors, UHDBT (type Qo-II) caused an increase of the anomalous signal of the FeS (Table 2 and Fig. 3C) and strong positive density for the ISP, for example, in the difference-density map between the UHDBT-bound and the native structures (Fig. 4). This density covered a region of the right shape and volume to fit the entire extramembrane domain of the ISP, indicating that the whole domain underwent a transition to a less mobile or completely immobilized state, in which it is bound to the surface of cytochrome b.
Figure 4.
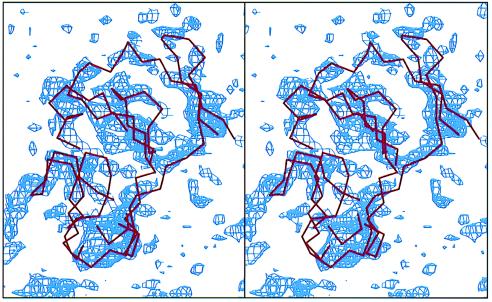
Stereo drawing: Difference-density map (blue) between the UHDBT-bound and native structures, contoured at the 2-σ level. Overlaid is the Cα trace (red) of the extramembrane domain of the ISP (22).
Binding of stigmatellin (type Qo-III) had an immobilizing effect on the ISP domain similar to that caused by UHDBT. Stigmatellin, which binds more strongly than UHDBT to most bc1 complexes (30), caused a larger increase in the height of the anomalous peak for the FeS than did UHDBT (Table 2).
ISP Extramembrane Domain.
In the crystal structure of native bc1 complex, the parts protruding into the intermembrane space were represented by weak, uninterpretable electron density (16). We used the position of the FeS obtained from anomalous scattering maps and the high-resolution model of the extramembrane domain of the ISP (22) in a rigid body search in real space to determine the orientation of the ISP domain. For this purpose we calculated electron density maps with data from crystals of native bc1 complex and of cocrystals with inhibitors. We used model phases to 3-Å resolution after cyclic solvent flattening; the starting phases did not contain contributions from the ISP extramembrane domain, and the solvent mask excluded an appropriate region around the iron position from flattening. Table 3 summarizes the results of our rigid body searches. Clearly, the orientation of the ISP domain was defined best for complexes containing the inhibitors of types Qo-II and Qo-III, with the correlation for stigmatellin slightly higher than for UHDBT. Fig. 5 shows the ISP domain in the orientation with optimal correlation; it contacts the extramembrane surface of cytochrome b. The correlation was significantly weaker in the native structure and very weak for myxothiazol and MOA-stilbene. A search around the possible alternative iron position in MOA-stilbene gave a very weak correlation, demonstrating that the structure of the ISP domain is not well defined in these crystals. The results of these searches support our interpretation that the decrease in the heights of the anomalous FeS peaks indicates a decreasing occupancy of the ISP domain bound to the surface of cytochrome b, with the occupancy strongest in the presence of stigmatellin and weakest in the presence of MOA-stilbene.
Table 3.
Constrained real space rotational and translational search with an extramembrane fragment of ISP as a search model
| Maps used in searches | Angular rotation,* ° | Changes in distance relative to native, Å | Maximum c.c. (×1000) | σ above mean, n | Second-largest c.c. (×1000) | σ above mean, n |
|---|---|---|---|---|---|---|
| Native | 0.0 | 0.0 | 186.6 | 11.4 | 68.1 | 4.1 |
| UHDBT | 3.3 | 0.7 | 313.1 | 15.5 | 84.9 | 3.8 |
| Stigmatallin | 4.2 | 1.1 | 309.8 | 17.6 | 76.3 | 3.9 |
| Myxothiazol | 10.6 | 1.2 | 102.1 | 5.9 | 67.7 | 3.9 |
| MOAS† | 134.2 | 15.1 | 73.1 | 4.7 | 61.6 | 3.9 |
c.c., Linear correlation coefficient.
Angular rotation is defined as a rotation that brings the position of ISP in inhibitor bound forms to the native position.
Putative new position of ISP in MOA-stilbene-bound bc1 crystal.
DISCUSSION
We have shown that different types of inhibitors of quinol oxidation by the cytochrome bc1 complex prefer different subsites within the Qo binding site in the cytochrome b subunit. This preference correlates well with the known spectroscopic effects of the Qo inhibitors on heme bL and/or on the FeS; it is not affected by the presence or absence of the Qi inhibitor antimycin A. Binding of Qo inhibitors has a dramatic effect on the conformational state of the ISP’s extramembrane domain: synonymous changes in the relative heights of FeS peaks in anomalous difference maps and in the correlations of a model density of the extramembrane ISP domain with unbiased electron densities in the various cocrystals indicate a large change in mobility of this domain. The bc1 crystals used in our studies are particularly suitable for observing such changes because the protein domains located in the mitochondrial intermembrane space–i.e., the extramembrane parts of cytochrome c1 and the ISP, and subunit 8—do not participate in crystal contacts, and can therefore undergo free movements or conformational transitions.
In crystals of native bc1 (Qo binding pocket empty), the ISP extramembrane domains appear to be partitioned between a fixed state and a loose state. Only the molecules in the fixed state can be observed by crystallography; they are bound to the surface of cytochrome b, above the surface helices cd1 and cd2 (Fig. 5). The FeS in the fixed state is 31 Å away from the iron in cytochrome c1; this distance makes fast ET between the FeS and the c1 heme unlikely.
Binding of myxothiazol or MOA-stilbene [type Qo-I (7)] shifts the equilibrium toward the loose state, as indicated by decreased anomalous FeS peak heights and density correlations. MOA-stilbene binding even leads to the complete disappearance of fixed ISP molecules and to the appearance of a new peak in the anomalous difference map, 15 Å away from the native position. This peak and a weak density correlation could indicate an alternative conformation of the ISP, in which the ISP would make close contact with cytochrome c1, rotated by 134° relative to the native orientation. However, because of the weakness of the density correlation and of the iron signal from anomalous scattering, we have to assume that this state, if it exists, is very poorly populated.
In contrast, the binding of UHDBT (type Qo-II) and stigmatellin (type Qo-III) strongly increases the population of the fixed ISP state. These inhibitors, although making intimate contacts with cytochrome b, also directly contact the ISP near the FeS. A close contact between stigmatellin and FeS was expected because of the large increase of the FeS’s redox potential upon inhibitor binding (11, 25). The detailed molecular mechanism of the transition of the ISP extramembrane domain from a fixed to a loose state in response to binding of ligands in the Qo site must await the completion of crystallographic refinement of the atomic model and the collection of crystallographic data on quinone binding at the Qo site.
One of the most important features of the Q cycle hypothesis is the bifurcation of ET at the Qo site, the mechanism of which is still under investigation. The transition between a fixed and a loose state of the ISP extramembrane domain can be integrated into a functional model of quinol oxidation very similar to the catalytic switch model proposed by Brandt and von Jagow (12). This model proposes that, upon reduction of FeS, the Qo center switches from an “FeS-state” that allows ET from the Qo site only to FeS, to a “b-state” that allows ET only to heme bL. The “b-state” in this model corresponds well to the loose state, whereas the “FeS-state” resembles the fixed state.
To combine our observations with the catalytic switch model, we assume two transient quinone binding sites in the inhibitor binding pocket. One of these putative sites, designated P1, is closer to ISP where UHDBT binds; the other site, designated P2, is closer to bL heme where MOA-stilbene binds. Analogous to inhibitor binding, binding of quinol to P1 will cause the fixation of ISP, and binding of semiquinone to P2 will release the ISP from this fixed state. With these assumptions, the ET events at the Qo site can be described as follows: A cycle starts with ubiquinol bound to P1, and the oxidized ISP in the fixed state. The first electron is transferred from ubiquinol to the FeS; the two protons are released. The ubisemiquinone moves to P2, causing a conformational transition in which the ISP is in the loose state. The reduced ISP in the loose state approaches cytochrome c1 and may form a transient complex with it; this allows rapid ET from the FeS to the c1 heme. The second electron is transferred from ubisemiquinone in P2 to heme bL, and subsequently to bH and to ubiquinone or ubisemiquinone in the Qi site. The ubiquinone in P2 dissociates from the bc1 complex to be replaced by another ubiquinol molecule, which will occupy P1 and bring the ISP back to the fixed conformational state.
One problem with this model is that especially Qo-I inhibitors are chemically dissimilar to quinones, so that it is not clear whether ubisemiquinone binds in P2, and, if it does, whether it can cause the same structural changes as MOA-stilbene. Another problem is the lack of a detectable ubisemiquinone radical at the Qo site. The reported detection of a transient, antimycin A-insensitive, ubisemiquinone radical at the Qo site (31) was recently questioned (P. R. Rich, personal communication), as it was not sensitive to the Qo site inhibitors such as myxothiazol, MOA-stilbene, or stigmatellin. An alternative hypothesis for the ET event at the Qo site could avoid this difficulty by assuming that the two electrons of ubiquinol in the complex are transferred simultaneously, one to FeS, and the second to heme bL and further to heme bH; thus, no ubisemiquinone would be generated. In this scenario, movement of the quinone between Qo subsites would probably not occur, and the conformational change of the cytochrome b protein, which switches the reduced ISP from the fixed to the loose state, would have a different cause. An attractive candidate for the switching event would be the ET from heme bL to heme bH; this mechanism would make sure that the second electron of ubiquinol would reach its destination before or at the same time as the first electron reaches heme c1. Thus, because the oxidation of the reduced FeS would depend on the transfer of an electron from heme bL to heme bH, bifurcation would be obligatory for the oxidation of ubiquinol in the bc1 complex.
During the preparation of this manuscript, Zhang et al. (32) reported the x-ray structure analysis of cytochrome bc1 complexes from chicken, beef, and rabbit; the four crystal forms described by these authors are all different from the one used in our work. One of the main findings was variability in the orientation of the extramembrane domain of the ISP in different crystal forms. In the presence of stigmatellin, the FeS position in the chicken bc1 complex appears identical to the one in the fixed state reported here. In the absence of stigmatellin, the FeS was found closer to the cytochrome c1 heme, with the ISP domain apparently in alternative fixed states. At least in some of the crystal forms the ISP in these states seems to be involved in, and stabilized by, crystal contacts (32); this would also explain the higher crystalline order of cytochrome c1 and subunit 8 (“hinge”) in these crystals. Nevertheless, the conclusion of Zhang et al. that the ISP extramembrane domain of the bc1 complex is mobile, and that its mobility has functional implications for ET, is identical to the conclusion we reached on the basis of our results.
Acknowledgments
We thank Dr. Stephen R. Sprang for thoughtful comments on the manuscript, Ms. Dorothee B. Staber for help with the manuscript, and the staff at beamlines X4A, X12B, and X25 at the National Synchrotron Light Source, BL-4 at the European Synchrotron Radiation Facility, and 7-1 at the Stanford Synchrotron Radiation Laboratory for help with data collection. This work was supported by National Institutes of Health Grant GM 30721 to C.-A.Y. and by a grant from the Welch foundation to J.D. J.D. is an Investigator in the Howard Hughes Medical Institute.
ABBREVIATIONS
- ET
electron transfer
- ISP
iron–sulfur protein
- FeS
iron–sulfur center
- MOA
methoxyacrylate
- UHDBT
5-undecyl-6-hydroxy-4,7-dioxobenzothiazol
References
- 1.Trumpower B L, Gennis R B. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 2.Brandt U, Trumpower B L. CRC Crit Rev Biochem Mol Biol. 1994;29:165–197. doi: 10.3109/10409239409086800. [DOI] [PubMed] [Google Scholar]
- 3.Trumpower B L. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatefi Y. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P. J Theor Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 6.Trumpower B L. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 7.von Jagow G, Link T A. Methods Enzymol. 1986;126:253–271. doi: 10.1016/s0076-6879(86)26026-7. [DOI] [PubMed] [Google Scholar]
- 8.Link T A, Haase U, Brandt U, von Jagow G. J Bioenerg Biomembr. 1993;25:221–232. doi: 10.1007/BF00762584. [DOI] [PubMed] [Google Scholar]
- 9.Brandt U, Schägger H, von Jagow G. Eur J Biochem. 1988;173:499–506. doi: 10.1111/j.1432-1033.1988.tb14026.x. [DOI] [PubMed] [Google Scholar]
- 10.Bowyer J R, Edwards C A, Ohnishi T, Trumpower B L. J Biol Chem. 1982;257:8321–8330. [PubMed] [Google Scholar]
- 11.Ohnishi T, Brandt U, von Jagow G. Eur J Biochem. 1988;176:385–389. doi: 10.1111/j.1432-1033.1988.tb14293.x. [DOI] [PubMed] [Google Scholar]
- 12.Brandt U, von Jagow G. Eur J Biochem. 1991;195:163–170. doi: 10.1111/j.1432-1033.1991.tb15690.x. [DOI] [PubMed] [Google Scholar]
- 13.Ding H, Robertson D E, Daldal F, Dutton P L. Biochemistry. 1992;31:3144–3158. doi: 10.1021/bi00127a015. [DOI] [PubMed] [Google Scholar]
- 14.Ding H, Moser C C, Robertson D E, Tokito M K, Daldal F, Dutton P L. Biochemistry. 1995;34:15979–15996. doi: 10.1021/bi00049a012. [DOI] [PubMed] [Google Scholar]
- 15.Brandt U. Biochim Biophys Acta. 1996;1275:41–46. doi: 10.1016/0005-2728(96)00048-5. [DOI] [PubMed] [Google Scholar]
- 16.Xia D, Yu C-A, Kim H, Xia J Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C-A, Yu L. Biochim Biophys Acta. 1980;591:409–420. doi: 10.1016/0005-2728(80)90172-3. [DOI] [PubMed] [Google Scholar]
- 18.Yu C-A, Xia J-Z, Kachurin A M, Yu L, Xia D, Kim H, Deisenhofer J. Biochim Biophys Acta. 1996;1275:47–53. doi: 10.1016/0005-2728(96)00049-7. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computing Project No. 4. A Suite of Programs for Protein Crystallography. Warrington, U.K.: Daresbury Laboratory; 1979. [Google Scholar]
- 21.Cowtan K D, Main P. Acta Crystallogr D. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- 22.Iwata S, Saynovits M, Link T A, Michel H. Structure. 1996;4:567–579. doi: 10.1016/s0969-2126(96)00062-7. [DOI] [PubMed] [Google Scholar]
- 23.Ohnishi T, Trumpower B L. J Biol Chem. 1980;255:3278–3284. [PubMed] [Google Scholar]
- 24.von Jagow G, Ljungdahl P O, Graf P, Ohnishi T, Trumpower B L. J Biol Chem. 1984;259:6318–6326. [PubMed] [Google Scholar]
- 25.von Jagow G, Ohnishi T. FEBS Lett. 1985;185:311–315. doi: 10.1016/0014-5793(85)80929-7. [DOI] [PubMed] [Google Scholar]
- 26.Geier B M, Schägger H, Brandt U, Colson A-M, von Jagow G. Eur J Biochem. 1992;208:375–380. doi: 10.1111/j.1432-1033.1992.tb17197.x. [DOI] [PubMed] [Google Scholar]
- 27.Robertson D E, Daldal F, Dutton P L. Biochemistry. 1990;29:11249–11260. doi: 10.1021/bi00503a014. [DOI] [PubMed] [Google Scholar]
- 28.Tron T, Crimi M, Colson A-M, Degli Esposti M. Eur J Biochem. 1991;199:753–760. doi: 10.1111/j.1432-1033.1991.tb16180.x. [DOI] [PubMed] [Google Scholar]
- 29.Tron T, Lemesle-Meunier D. Curr Genet. 1990;18:413–419. doi: 10.1007/BF00309910. [DOI] [PubMed] [Google Scholar]
- 30.Geier B M, Haase U, von Jagow G. Biochem Soc Trans. 1994;22:203–209. doi: 10.1042/bst0220203. [DOI] [PubMed] [Google Scholar]
- 31.de Vries S, Albracht S P J, Berden J A, Slater E C. Biochim Biophys Acta. 1982;681:41–53. doi: 10.1016/0005-2728(82)90276-6. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z L, Huang L S, Shulmeister V M, Chi Y I, Kim K K, Hung L W, Crofts A R, Berry E A, Kim S H. Nature (London) 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 34.Evans S V. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]