Abstract
Specific mutations in BRCA1 and BRCA2 genes have been identified in specific populations and ethnic groups. However, little is known about the contribution of BRCA1 and BRCA2 mutations to breast cancers in the Indonesian population. One hundred-twenty moderate to high risk breast cancer patients were tested using PCR-DGGE, and any aberrant band was sequenced. Multiplex ligation-dependent probe amplification (MLPA) was performed on all samples to detect large deletions in the two genes. Twenty-three different mutations were detected in 30 individuals, ten were deleterious mutations and 20 were “unclassified variants” with uncertain clinical consequences. Three of seven (c.2784_2875insT, p.Leu1415X and del exon 13–15) and two of four (p.Glu2183X and p.Gln2894X) deleterious mutations that were found in BRCA1 and BRCA2 respectively, are novel. Several novel, pathogenic BRCA1 and BRCA2 germline mutations are found in early onset Indonesian breast cancer patients, these may therefore be specific for the Indonesian population.
Keywords: BRCA1, BRCA2, Hereditary breast cancer, Mutation analysis
Introduction
Breast cancer is the most common cancer in women. In 5% to 10% of breast cancer cases, the disease results from a hereditary predisposition [1, 2], which can to a large extent be attributed to mutations in either of two tumour suppressor genes, BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) [3–5]. These genes are involved in DNA repair as well as transcriptional regulation [6, 7]. Women carrying pathogenic germline mutations in either of these genes tend to develop breast cancer at an early age [8, 9].
The BRCA1 and BRCA2 genes encode large proteins of 1,863 and 3,418 amino acids, respectively. Over 300 distinct mutations in BRCA1 and BRCA2 have been described [10, 11]. These mutations are widely scattered across both genes and most affect the structure and function of the gene. Nevertheless, a significant proportion (34% of BRCA1 and 38% of BRCA2 mutations) (http://www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic) are missense mutations that alter one amino acid, but do not truncate the protein and are rare sequence variants of unknown functional consequence. Moreover, a number of base substitutions do not alter the amino acid sequence or result in amino acid changes not associated with disease (polymorphisms) [12]. Hence the biggest challenge in interpreting the mutation analysis of BRCA1 and BRCA2 genes is to distinguish between harmless polymorphisms and deleterious mutations associated with increased cancer risk.
In addition, mutations specific for certain populations and ethnic groups have been identified in both genes. For example, specific BRCA1 and BRCA2 mutations were reported for Ashkenazi Jews [13]. Other common BRCA1 mutations were especially found in Italian, Canadian, Belgian or Dutch breast cancer families [14–16]. In Indonesia, the contribution of the BRCA1/BRCA2 mutations to the population incidence of early-onset breast cancer is largely unknown. In one pilot study, however, a new BRCA2 mutation was identified [17] indicating that it was worthwhile to more extensively study the Indonesian population, which was the aim of this study. The accumulating knowledge about the prevalence and nature of BRCA1 and BRCA2 mutations in specific populations may facilitate the interpretation of genetic analysis with regard to breast cancer risk of individual patients.
Materials and methods
Patients
A total of 120 unrelated breast cancer patients and 16 of their family members from three Indonesian cities (Jakarta and Jogjakarta on the Java island, Denpasar on the Bali island) were analyzed. Breast cancer patients at moderate to high-risk of a hereditary predisposition were selected according to the following criteria: (A) Breast cancer before the age of 41 (n = 102); (B) Two cases of breast cancer in the same family before the age of 60 (n = 9); (C) Three or more cases of breast cancer in the same family (n = 2); (D) Bilateral breast cancer (n = 7). Subjects were asked to fill out questionnaires to evaluate their personal and family histories, and blood specimens were collected for determination of BRCA mutations. Informed consent was obtained from all the subjects in this study.
DNA extraction and PCR amplification
Genomic DNA was isolated by the saturated salt extraction procedure as described in [18]. All 22 coding exons of BRCA1 and 26 coding exons of BRCA2 were amplified using primer sequences developed by the University of Groningen, The Netherlands [19]. Primers for DGGE were obtained from Ingeny (Goes, The Netherlands). Genomic DNA was amplified using 100–200 ng of template DNA, 10 pmol of the mixture of 40-mer primers, 30 mM of MgCl, 3 mM dNTPs (Invitrogen) and 0.7 unit of Platinum Taq (Invitrogen) in 9 μl PCR reactions. In order to speed up the test, the PCR reaction was placed in 384 well plates using a pipetting robot (TECAN Miniprep 75). PCR conditions were performed as previously described [17].
Denaturing Gradient Gel Electrophoresis and DNA sequencing
A 4–6 μl aliquot of each PCR product with relatively large melting temperature differences were pooled as previously described [17] with some modifications for optimal results. The fragment pool was designed based on melting profiles and sequence. Electrophoresis was performed in 0.5 TAE buffer at 58°C, 120 V for 16 h for BRCA1 gene, and 55°C, 100 V for 18 h for BRCA2. Gels were stained with ethidium bromide and photographed under a UV transilluminator. The aberrantly migrating samples were re-amplified using sequencing primers and sequencing was performed using Big Dye Cycle-sequencing kit according to the manufacturer’s instructions. The reaction products were analyzed using an ABI 3100 DNA Sequencer (Applied Biosystems, Torrence, CA, USA) and sequence files were edited using the Bio Edit program. The classification of gene alterations was performed in accordance with the entries in the Breast Cancer Information Core (BIC, Bethesda, MD).
Multiplex ligation-dependent probe amplification (MLPA)
The principle of the MLPA technique has been described elsewhere [20]. The MLPA test for BRCA1 (P02) and BRCA2 (P45) mutations were obtained from MRC-Holland, Amsterdam, The Netherlands. The fragments were analyzed on an ABI model 310 capillary sequencer (Applied Biosystems, Torrence, CA, USA) using Genescan-TAMRA 500 size standards (Applied Biosystems). Fragment analysis was performed with Genescan software.
Results and discussions
We identified 120 incident Indonesian breast cancer cases diagnosed before the age of 41 years, or having family history of breast cancer, or harboring bilateral breast cancer during September 1999–April 2005 (Jogjakarta) and during July 2004–April 2005 (Jakarta and Denpasar). In addition, 16 of their family members were analyzed.
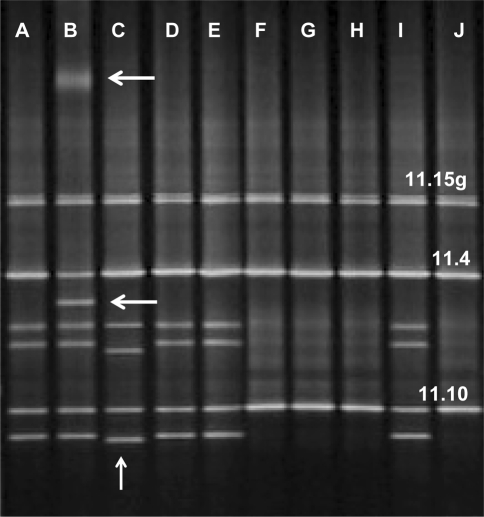
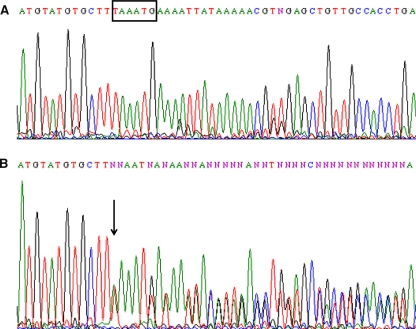
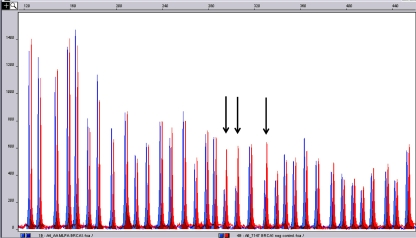
The entire coding regions and exon-intron junctions of BRCA1 and BRCA2 were screened in these 136 persons of breast cancer patients and their families using PCR-DGGE (Fig. 1) followed by sequencing (Fig. 2) for samples with aberrant migrating bands. To optimize the screening, MLPA, a relatively new technique, was also performed in all samples (Fig. 3). Here, we report on 116/120 women (96.7%) for whom BRCA1/2 analysis were completed. The remaining four patients (all from group A) had to be excluded due to the small amount of extracted DNA that did not allow complete screening of the BRCA1 and BRCA2 genes.
Fig. 1.
DGGE analysis of fragments 11.15 g, 11.4 and 11.10 of the BRCA2 gene in ten unrelated breast cancer patients. The arrows show altered band mobility compare to other patients
Fig. 2.
Sequence electropherogram of a normal individual showing (A) wild-type BRCA2 exon 11 sequence and (B) of breast cancer patient (B-3-5) showing c.2699_2704delTAAATG mutation
Fig. 3.
MLPA analysis of BRCA1 gene of patient sample (blue) compare to the normal control (red). X and Y axis represent peak size and peak height respectively. There are reduced peaks in the patient sample compared to the normal control in exons 13, 14 and 15 (arrows) indicating deletions
BRCA1 and BRCA2 pathogenic mutations
The analysis of 116 unrelated breast cancer patients with breast cancer revealed that nine patients (7.8%) carried pathogenic germline mutations especially the early onset patients: 3 within BRCA1 (2.6%) and 6 within BRCA2 (5.2%) which is comparable to previous studies [21]. We only found BRCA1 and BRCA2 mutations in groups A (“early onset”, n = 7 out of 98, 7.1%) and B (two cases of breast cancer in the same family before the age of 60, n = 2 out of 9 (22.2%)) (Table 1). There were twice as many BRCA2 mutations as BRCA1 mutations. Although the absolute numbers are low and no firm conclusions can therefore be drawn, this is comparable to other Asian regions [22–24] but seems to discern the Indonesian population from non-Asian ethnic groups where the reverse trend is seen.
Table 1.
BRCA1 or BRCA2 germline mutations in Indonesian women with early onset breast cancer
| Patient | Agea | gene | Exon | Mutationb | mutation type | Pathogenic mutation | BICc |
|---|---|---|---|---|---|---|---|
| AE | 25 | BRCA1 | 11 | c.2784_2785insT | frameshift | + | no |
| B10 | 31 | BRCA1 | 13 | p.Leu1415X | nonsense | + | no |
| AA | 40 | BRCA1 | 13–15 | −d | large rearrangement | + | no |
| AB | 34 | BRCA2 | 11 | c.3040_3043del4 | frameshift | + | 1 |
| B5 | 66 | BRCA2 | 11 | p.Glu2183X | nonsense | + | no |
| B6 | 65 | BRCA2 | 11 | p.Glu2183X | nonsense | + | no |
| B-III-5 | 30 | BRCA2 | 11 | p.Leu824X | nonsense | + | no |
| AZ | 40 | BRCA2 | 11 | p.Leu824X | nonsense | + | no |
| W-II | 37 | BRCA2 | 21 | p.Gln2894X | nonsense | + | no |
| Q-II | 40 | BRCA1 | 2 | c.101–10T>C | IVS | ± | 6 |
| P-III-19 | 19 | BRCA1 | 9 | p.Val191Ile | Missense | ± | 6 |
| J22 | 32 | BRCA1 | 11 | p.Leu1209Val | Missense | ? | no |
| AZ | 40 | BRCA1 | 16 | p.Met1652Ile | Missense | ± | 35 |
| B1 | 24 | BRCA1 | 20 | c.5313–31A>G | IVS | ? | no |
| B7 | 31 | BRCA1 | 24 | p.Arg1835Gln | Missense | ? | no |
| 216 | 33 | BRCA1 | 24 | p.Thr1852Ile | Missense | ? | no |
| P-III-19 | 19 | BRCA2 | 5 | p.Gln147Arg | Missense | ± | 6 |
| B3 | 24 | BRCA2 | 10 | p.Gln609Glu | Missense | ? | no |
| C-II-7 | 39 | BRCA2 | 11 | p.Met1149Val | Missense | ± | 5 |
| AO | 28 | BRCA2 | 11 | p.Met1149Val | Missense | ± | 5 |
| AQ | 44 | BRCA2 | 11 | p.Met1149Val | Missense | ± | 5 |
| BH | 38 | BRCA2 | 11 | p.Met1149Val | Missense | ± | 5 |
| 172 | 36 | BRCA2 | 11 | p.Gln699Leu | Missense | ? | no |
| J32 | 29 | BRCA2 | 11 | p.Arg2108Cys | Missense | ± | 16 |
| J6 | 33 | BRCA2 | 11 | p.Val950Ile | Missense | ? | no |
| 206 | 37 | BRCA2 | 25 | c.9485–16T>C | IVS | ± | 4 |
| BC | 35 | BRCA2 | 27 | p.Ile3412Val | Missense | ± | 109 |
| 166 | 33 | BRCA2 | 27 | p.Ile3412Val | Missense | ± | 109 |
| J24 | 35 | BRCA2 | 27 | p.Ile3412Val | Missense | ± | 109 |
| 206 | 37 | BRCA2 | 27 | p.Lys3326X | nonsense | ± | 289 |
Seven pathogenic mutations were found in nine probands: three in BRCA1 (c.2784_2785insT, pL1415X (c.4361_4362insT), del exon 13–15) and four in BRCA2 (c.3040_3043delGCAA, p.Glu2183X (c.6775G>T), p.Leu824X (c.2699_2704delTAAATG), p.Gln2894X (c.9008C>T)). All these mutations were classified as pathogenic as they are predicted to result in protein truncation. The three pathogenic mutations found in BRCA1 were not previously reported in the BIC database as well as two novel nonsense mutations (p.Glu2183X and p.Gln2894X) identified in BRCA2. The p.Glu2183X mutation was found in 2 related patients that had breast cancer above the age of 60.
One of seven pathogenic mutations found in BRCA1 and BRCA2 showed a significant clinical impact on the patient (Table 2). Patient AE with a one nucleotide insertion (Thymine) between nucleotide 2784 and 2785 (c.2784_2785insT) in exon 11 of BRCA1 suffered from bilateral breast cancer at a relatively early age (25 years). The insertion leads to frameshift and creates a premature stop codon in exon 11. The mutation takes place in the sequence within BRCA1 encoding for aminoacids 758–1064 which interact with RAD51 protein that is required for homologous recombination (HR) repair of double strand breaks (DSBs) [25], which is one of the most important functions of the BRCA1 protein. This patient presented in a late stage (stage III for both breasts) and only survived for 9 weeks after treatment. Her mother did not carry this mutation. Although her father may be carrier, the mutation is probably de novo as there was no family history of breast or other cancers.
Table 2.
Clinicopathological features of Indonesian breast cancer patients with deleterious BRCA1 or BRCA2 germline mutations
| Patient | Agea | Gene with germline mutation | Mutationb | stage | Diagnosis | Menopausal status | family history of cancer | Survival status |
|---|---|---|---|---|---|---|---|---|
| AE | 25 | BRCA1 | c.2784_2785insT | IIIB/IIIA | IDC, bilateral | pre | No | DOD 9 w |
| B10 | 31 | BRCA1 | p.Leu1415X | I | IDC | pre | No | DOD 57 w |
| AA | 40 | BRCA1 | -c | IIIB | IDC N+ | pre | No | AWD |
| AB | 34 | BRCA2 | c.3040_3043del4 | IIIB | IDC N+ | pre | Sister, Int | DOD 17 w |
| B5 | 63 | BRCA2 | p.Glu2183X | IV | Tubular | post | Sister,Br | AWD |
| B6 | 65 | BRCA2 | p.Glu2183X | III | IDC | post | Brother, Br | AWD |
| B-III-5 | 30 | BRCA2 | p.Leu824X | I | IDC | pre | No | AWD |
| AZ | 40 | BRCA2 | p.Leu824X | IV | IDC | pre | Sister, Cv | DOD 46 w |
| W-II | 37 | BRCA2 | p.Gln2894X | IIIA | IDC | pre | No | DOD 107 w |
The second pathogenic mutation with a significant clinical manifestation was a cytosine for thymine substitution on nucleotide 9008 of BRCA2 leading to a premature stop codon in position 2894, c9008C>T (p.Gln2894X). Patient W presented at age 37 in a late stage and survived for only 107 weeks after initial treatment. She had no family history of breast or other cancers. This mutation lies within exon 21 of BRCA2 which is the proposed site for interaction with the DSS1 protein that seems to have a fundamental role in enabling the BRCA2-RAD51 complex to associate with sites of DNA damage [26].
The c.2699_2704delTAAATG (p.Leu824X) in BRCA2 that has been reported previously by us in the Indonesian population [17], was found in one other patient in the present study (Table 1). This mutation lies in exon 11 BRCA2, within the BRC repeats domain. The truncating mutation causes loss of three quarters of the protein leading to lack of interaction with the RAD51 protein. Different from BRCA1, the repair of DSBs by HR is the most important function of the BRCA2 protein [27]. Patient B-III-5 was diagnosed with early stage breast cancer at age 30 with no family history of breast or other cancers. Her sister carried the same mutation, but with no present clinical manifestation as yet. Patient AZ who was diagnosed at 40 years of age, presented in late stage, only survived 46 weeks after initial treatment. This patient also harbored a mutation in exon 16 of BRCA1, a G to A substitution in nucleotide 5075 (c.5075G>A), which leads to amino acid change from Methionine to Isoleucine, (p.Met16521Ile) which has to date been reported 35 times in BIC as a UV mutation. As the c.2699_2704delTAAATG mutation was found in two unrelated patients, this mutation could be a good candidate as a founder mutation.
None of the families with more than 3 cases of breast cancer and families with bilateral breast cancer showed pathogenic mutations in the BRCA1 and BRCA2 genes. Family U had four first-degree relatives that were affected by breast cancer. Two of four members had bilateral breast cancer. In spite of this high familial breast cancer incidence, no BRCA1/2 mutations were found.
BRCA1 and BRCA2 unclassified variants
Sixteen (7 BRCA1 and 9 BRCA2) rare mutations of so far unknown significance (“unclassified variants”, UVs) were detected in 18 patients: 13 missense changes and 3 intronic variants. Of these 16 UVs, 7 were novel, whereas the other UVs have been previously reported in the BIC database (Table 1). From the 18 patients which carried UV mutations, two patients were detected in families from group D; one patient in a group B family and the other fifteen patients in families from group A.
Seven UV were found in the BRCA1 gene, two mutations occurring in the intronic region between exons 1 and 2 (c.101–10T>C) and between exons 19 and 20 (c.5313–31A>G), and five missense mutations identified: p.Val191Ile (c.690G>A), p.Leu1209Val (c.3744T>G), p.Met1652Ile (c.5075G>A), p.Arg1835Gln (c.5623G>A) and p.Thr1852Ile (c.5674C>T).
Four out of seven BRCA1 missense mutations; p.Leu1209Val (c.3744T>G), c.5313–31A>G, p.Arg1835Gln (c.5623G>A) and p.Thr1852Ile (c.5674C>T) were have not been described previously in the BIC. The p.Leu1209Val may not be a significant change as both Leucine and Valine belong to the same group of non polar amino acids. However, p.Arg1835Gln is possibly an important alteration since a positively charged Arginine is replaced by an uncharged Glutamine, which may have an effect on the structure and/or function of the protein. Another potentially important alteration concerns p.Thr1852Ile, where the hydrophilic amino acid Threonine is replaced by a hydrophobic Isoleucine. The sites of mutation of both p.Arg1835Gln and p.Thr1852Ile also have to be considered as they lie within the site for the activation domain of the BRCA1 protein [28]. The intronic UV c.5313–31A>G also deserves further investigation as it may theoretically have an effect on splicing. However, according to splice site finder (http://www.genet.sickkids.on.ca/∼ali/splicesitefinder.html), the splicing sites in the wild type and mutant alleles are similar, so therefore we can suggest that the c.5313–31A>G has no effect on splicing.
Nine different UVs of the BRCA2 gene were found in fourteen patients (Table1), and three of them were novel; p.Gln609Glu 9c.2053C>G), p.Gln699Leu (2324A>T) and p.Val950Ile (3076G>A). One truncating mutation near the C-terminal end of BRCA2, p.Lys3326X (c.10204A>T) is probably not pathogenic. Since the truncating mutation is at the very end of the protein, it is possible that protein functions are not affected. Most of the few entries in databanks describing nonsense mutations near the C terminus of BRCA2 between codon 3308 and 3408 are described as UVs. Thus, the effect of this truncating mutation on cancer predisposition remains unclear.
The p.Val950Ile may not be a significant change as both Valine and Isoleucine belong to the same group of non polar, hydrophobic amino acids. However, p Gln609Glu and p.Gln699Leu are potentially important alterations as for p.Gln609Glu, a non acidic, polar, hydrophilic Glutamine is replaced by a negatively charged Glutamic acid, whereas for p.Gln699Leu, an uncharged hydrophilic Glutamine is replaced by a hydrophobic Leucine. As it takes place within the BRC repeats of the BRCA2 protein, the p Gln699Leu alteration might affect protein structure and function.
To know more about the importance of amino acid substitutions for protein function, we compared the amino acid sequence of interest in seven other species, i.e. Mus musculus, Rattus rattus, Bos taurus, Gallus gallus, Canis familiaris, Macaca mullata and Monodelphis domestica. The missense mutation p.Leu1209Val lies in the conserved region of exon 11 of the BRCA1 gene as the sequence is maintained in seven other species, whereas p.Arg1835Gln and p.Thr1852Ile are only conserved in four and three other species (comparison of p.Arg1835Gln and p.Thr1852Ile with Bos taurus sequence is not possible because the BRCA1 gene is shorter). Therefore, even tough the Leucine to Valine changes may not give any effect on amino acid charge, its conservation in evolution is suggestive of a functional role. Interestingly, p.Gln609Glu and p.Gln699Leu of BRCA2 that result in a quite dramatic amino acid subtitution that might lead to protein structure changes, are only conserved in four and five species respectively. As for the p.Val950Ile, the conservation in evolution is quite low. Although p.Gln609Glu is less conserved, we still believe that Glutamine to Glutamic acid substitution may have an effect on protein conformation as two adjacent acidic amino acids will be formed as the result of the substitution.
Glycosylation moiety of an amino acid also plays a role in protein function. Amino acid substitutions involving Serine, Threonine and Asparagine, should also be checked for their O-GlcNac potential and threshold. Here we have a Threonine to Isoleucine substitution (p.Thr1852Ile) that after checking with YinOYang (http://www.cbs.dtu.dk/services/YinOYang) showed no significant threshold changes between the wildtype and the mutant allele.
The possible effect of amino acid changes in proteins can also be assessed using similarity scores (based on Grantham table [29]), in which a value above 100 for an amino acid substitution indicates a higher chance of impact on protein function. Among seven novel UVs in the BRCA1 and BRCA2 genes found in the present study, only p.Gln699Leu in BRCA2 has a similarity score above 100, whereas p.Gln609Glu and p.Val905Ile in BRCA2 have the lowest score (Table 3).
Table 3.
The amino acid properties of novel unclassified mutations in BRCA1 and BRCA2 within an Indonesian breast cancer population
| Gene | Amino acid change | Change of charge | Change of amino acid group | Similarity scorea | # species with conserved sequence |
|---|---|---|---|---|---|
| BRCA1 | Leu to Val | None | No | 32 | 7a,b,c,d,e,f,g |
| BRCA1 | Arg to Gln | Pos to no charge | Yes | 43 | 4a,c,f,g |
| BRCA1 | Thr to Ile | polar to non polar | Yes | 89 | 3a,c,g |
| BRCA2 | Gln to Glu | No charge to neg | Yes | 29 | 4a,b,c,g |
| BRCA2 | Gln to Leu | Polar to non polar | Yes | 113 | 5a,b,d,e,f |
| BRCA2 | Val to Ile | None | No | 29 | 2f,g |
abased on Grantham table [Grantham et al. [29], a score above 100 indicates significance changes
a = Macaca mullata, b = Bos taurus, c = Canis familiaris, d = Rattus rattus, e = Mus musculus, f = Gallus gallus, h = Monodelphis domestica
Overall, we propose that among the seven novel UVs, there are three mutations that are possibly pathogenic: p.Leu1209Val for its location in a conserved region, and p.Gln609Glu and the p.Gln699Leu because of two adjacent acidic amino acid being formed and a high similarity score, respectively.
When comparing the three different Indonesian regions, the percentages of breast cancer patients with pathogenic BRCA1/2 mutations was significantly higher in Denpasar (Bali island) than in Jogjakarta and Jakarta (Java island) ((25% (3/12), 7.2% (6/83) and 0% (0/25) respectively (P = 0.0255, chi-square test)). The percentages of breast cancer patients with UV mutations in Jakarta, Jogjakarta, and Denpasar were 16% (4/25), 12% (10/83), and 25% (3/12), respectively (n.s.). Although the number of patients is too small to draw firm conclusions, these data may point to geographic differences within Indonesia.
It was initially suggested that the BRCA1 and BRCA2 genes would be responsible for most cases of inherited breast cancer, but more recent studies suggest that they would account for a far smaller proportion, with considerable variation among different populations [30]. We found that the incidence of mutations in these genes varies, depending on the diagnostic group. In this sense, mutations were present in (22/102) 21.6% of early onset patients (group A), 28.7% (2/7) in patients with bilateral breast cancer (group D) and (2/9) 22.2% of patients with two cases of breast cancer before the age of 60 (group B). The proportion of families affected by BRCA1/2 mutations depends on the population analyzed and on the criteria used to select the patients. Family history of breast cancer was, however, absent or not suggestive of a hereditary predisposition in three-fourth of the deleterious mutations carriers and in more than 90% of UV carriers. This suggests that BRCA screening policies based on family history only would miss a considerable proportion of mutation carriers.
In conclusion, a relatively high percentage of early onset Indonesian breast cancer patients carry a germline mutation in either BRCA1 or BRCA2. Several novel, pathogenic BRCA1 and BRCA2 germline mutations have been found, as well as a variety of novel “unclassified variant” mutations that may therefore be specific for the Indonesian population. It is likely that some of the “unclassified variant” mutations may have a functional role in breast cancer development, which deserves to be explored further.
Acknowledgements
Supported by grant IN-2001–008 of the Dutch Cancer Society. We thank dr. Jo Hilgers who has been instrumental in setting up the Familial Cancer Clinic initiative in Jogjakarta, and Jan Schouten for the technical support.
References
- 1.Easton D, Peto J (1990) The contribution of inherited predisposition to cancer incidence. Cancer Surv 9(3):395–416 [PubMed]
- 2.Claus EB, Schildkraut JM, Thompson WD, Risch NJ (1996) The genetic attributable risk of breast and ovarian cancer. Cancer 77(11):2318–2324 [DOI] [PubMed]
- 3.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250(4988):1684–1689 [DOI] [PubMed]
- 4.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266(5182):66–71 [DOI] [PubMed]
- 5.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378(6559):789–792 [DOI] [PubMed]
- 6.Scully R, Livingston DM (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408(6811):429–432 [DOI] [PMC free article] [PubMed]
- 7.Zheng L, Li S, Boyer TG, Lee WH (2000) Lessons learned from BRCA1 and BRCA2. Oncogene 19(53):6159–6175 [DOI] [PubMed]
- 8.Rahman N, Stratton MR (1998) The genetics of breast cancer susceptibility. Annu Rev Genet 32:95–121 [DOI] [PubMed]
- 9.Stratton MR, Wooster R (1996) Hereditary predisposition to breast cancer. Curr Opin Genet Dev 6(1):93–97 [DOI] [PubMed]
- 10.Shattuck-Eidens D, McClure M, Simard J, Labrie F, Narod S, Couch F, Hoskins K, Weber B, Castilla L, Erdos M et al (1995) A collaborative survey of 80 mutations in the BRCA1 breast and ovarian cancer susceptibility gene. Implications for presymptomatic testing and screening. JAMA 273(7):535–541 [DOI] [PubMed]
- 11.Shattuck-Eidens D, Oliphant A, McClure M, McBride C, Gupte J, Rubano T, Pruss D, Tavtigian SV, Teng DH, Adey N et al (1997) BRCA1 sequence analysis in women at high risk for susceptibility mutations. Risk factor analysis and implications for genetic testing. JAMA 278(15):1242–1250 [DOI] [PubMed]
- 12.Monteiro AN, August A, Hanafusa H (1997) Common BRCA1 variants and transcriptional activation. Am J Hum Genet 61(3):761–762 [DOI] [PMC free article] [PubMed]
- 13.Roa BB, Boyd AA, Volcik K, Richards CS (1996) Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat Genet 14(2):185–187 [DOI] [PubMed]
- 14.Simard J, Tonin P, Durocher F, Morgan K, Rommens J, Gingras S, Samson C, Leblanc JF, Belanger C, Dion F et al (1994) Common origins of BRCA1 mutations in Canadian breast and ovarian cancer families. Nat Genet 8(4):392–398 [DOI] [PubMed]
- 15.Peelen T, van Vliet M, Petrij-Bosch A, Mieremet R, Szabo C, van den Ouweland AM, Hogervorst F, Brohet R, Ligtenberg MJ, Teugels E et al (1997) A high proportion of novel mutations in BRCA1 with strong founder effects among Dutch and Belgian hereditary breast and ovarian cancer families. Am J Hum Genet 60(5):1041–1049 [PMC free article] [PubMed]
- 16.Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, Hageman S, Arts PJ, Ligtenberg MJ et al (1997) BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 17(3):341–345 [DOI] [PubMed]
- 17.Purnomosari D, Paramita DK, Aryandono T, Pals G, van Diest PJ (2005) A novel BRCA2 mutation in an Indonesian family found with a new, rapid, and sensitive mutation detection method based on pooled denaturing gradient gel electrophoresis and targeted sequencing. J Clin Pathol 58(5):493–499 [DOI] [PMC free article] [PubMed]
- 18.Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3):1215 [DOI] [PMC free article] [PubMed]
- 19.Hout AH, Ouweland AM, Luijt RB, Gille HJ, Bodmer D, Bruggenwirth H, Mulder IM, Vlies PV, Elfferich P, Huisman MT et al (2006) A DGGE system for comprehensive mutation screening of BRCA1 and BRCA2: application in a Dutch cancer clinic setting. Hum Mutat [DOI] [PubMed]
- 20.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30(12):e57 [DOI] [PMC free article] [PubMed]
- 21.de Sanjose S, Leone M, Berez V, Izquierdo A, Font R, Brunet JM, Louat T, Vilardell L, Borras J, Viladiu P et al (2003) Prevalence of BRCA1 and BRCA2 germline mutations in young breast cancer patients: a population-based study. Int J Cancer 106(4):588–593 [DOI] [PubMed]
- 22.Liede A, Narod SA (2002) Hereditary breast and ovarian cancer in Asia: genetic epidemiology of BRCA1 and BRCA2. Hum Mutat 20(6):413–424 [DOI] [PubMed]
- 23.Ho GH, Phang BH, Ng IS, Law HY, Soo KC, Ng EH (2000) Novel germline BRCA1 mutations detected in women in singapore who developed breast carcinoma before the age of 36 years. Cancer 89(4):811–816 [DOI] [PubMed]
- 24.Takeda M, Ishida T, Ohnuki K, Suzuki A, Sakayori M, Ishioka C, Nomizu T, Noguchi S, Matsubara Y, Ohuchi N (2004) Collaboration of breast cancer clinic and genetic counseling division for BRCA1 and BRCA2 mutation family in Japan. Breast Cancer 11(1):30–32 [DOI] [PubMed]
- 25.Shinohara A, Ogawa H, Ogawa T (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69(3):457–470 [DOI] [PubMed]
- 26.Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A (2004) DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep 5(10):989–993 [DOI] [PMC free article] [PubMed]
- 27.Rudkin TM, Foulkes WD (2005) BRCA2: breaks, mistakes and failed separations. Trends Mol Med 11(4):145–148 [DOI] [PubMed]
- 28.Somasundaram K (2003) Breast cancer gene 1 (BRCA1): role in cell cycle regulation and DNA repair–perhaps through transcription. J Cell Biochem 88(6):1084–1091 [DOI] [PubMed]
- 29.Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185(4154):862–864 [DOI] [PubMed]
- 30.Szabo CI, King MC (1997) Population genetics of BRCA1 and BRCA2. Am J Hum Genet 60(5):1013–1020 [PMC free article] [PubMed]