Abstract
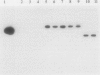
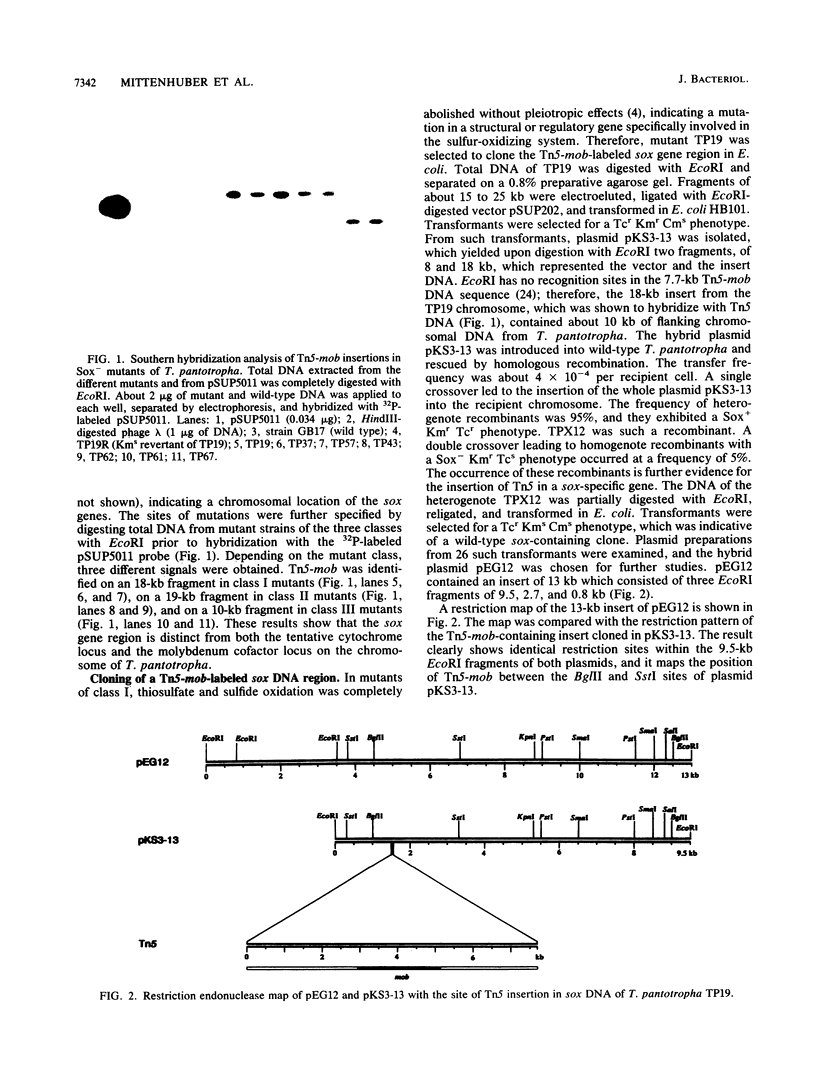
For the identification of the DNA region responsible for the sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha, we used previously isolated Tn5-mob insertional Sox- mutants. For seven mutants, the Tn5-mob insertion was localized on the chromosome rather than on the megaplasmids pHG41 or pHG42 by using the Tn5-mob-harboring vehicle pSUP5011 as probe. The specific insertion of Tn5-mob into a sox gene was determined for one Sox- mutant, strain TP19. An 18-kb EcoRI fragment was cloned in Escherichia coli by using the mobilizable plasmid pSUP202 as vector and the kanamycin resistance gene of Tn5 as marker. Conjugal transfer of the resulting hybrid plasmid, pKS3-13, to the wild type resulted in two phenotypically different groups of recombinants. Ninety-five percent of the recombinants were Sox+, kanamycin resistant, and tetracycline resistant; 5% were homogenote recombinants exhibiting the Sox-, kanamycin-resistant, tetracycline-sensitive phenotype, and these indicated the specific insertion. To isolate the respective wild-type sox gene, total DNA from a heterogenote recombinant was partially restricted with EcoRI, religated, and transformed in E. coli. Transformants carrying a pSUP202-derived hybrid plasmid with the intact sox gene were identified by screening for a tetracycline-resistant, kanamycin-sensitive, and chloramphenicol-sensitive phenotype and by complementation of the Sox- mutant TP19. A plasmid of this type, pEG12, contained an insert of 13 kb which gave a positive signal in Southern hybridization with the homologous probe of pKS3-13. pEG12 was used to determine the DNA homology of the sulfur-oxidizing enzyme systems of other thiobacteria. Strong hybridization signals were obtained with total DNA of the neutrophilic sulfur-oxidizing bacteria Paracoccus denitrificans, Thiobacillus versutus, and Rhodobacter capsulatus. No hybridization signal was obtained with DNA of other neutrophilic or acidophilic thiobacteria examined.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barros M. E., Rawlings D. E., Woods D. R. Cloning and expression of the Thiobacillus ferrooxidans glutamine synthetase gene in Escherichia coli. J Bacteriol. 1985 Dec;164(3):1386–1389. doi: 10.1128/jb.164.3.1386-1389.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chandra T. S., Friedrich C. G. Tn5-induced mutations affecting sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J Bacteriol. 1986 May;166(2):446–452. doi: 10.1128/jb.166.2.446-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobner E., Huber H., Stetter K. O. Thiobacillus ferrooxidans, a facultative hydrogen oxidizer. Appl Environ Microbiol. 1990 Sep;56(9):2922–2923. doi: 10.1128/aem.56.9.2922-2923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Hogrefe C., Schlegel H. G. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J Bacteriol. 1981 Jul;147(1):198–205. doi: 10.1128/jb.147.1.198-205.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay R., Silver M. Thiobacillus acidophilus sp. nov.; isolation and some physiological characteristics. Can J Microbiol. 1975 Mar;21(3):281–288. doi: 10.1139/m75-040. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Lobos J. H., Bopp L. H., Welch G. C. Cloning of a Thiobacillus ferrooxidans plasmid in Escherichia coli. J Bacteriol. 1984 Jan;157(1):324–326. doi: 10.1128/jb.157.1.324-326.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue C., Sugawara K., Kusano T. Thiobacillus ferrooxidans mer operon: sequence analysis of the promoter and adjacent genes. Gene. 1990 Nov 30;96(1):115–120. doi: 10.1016/0378-1119(90)90349-v. [DOI] [PubMed] [Google Scholar]
- Pretorius I. M., Rawlings D. E., Woods D. R. Identification and cloning of Thiobacillus ferrooxidans structural nif genes in Escherichia coli. Gene. 1986;45(1):59–65. doi: 10.1016/0378-1119(86)90132-0. [DOI] [PubMed] [Google Scholar]
- Römermann D., Warrelmann J., Bender R. A., Friedrich B. An rpoN-like gene of Alcaligenes eutrophus and Pseudomonas facilis controls expression of diverse metabolic pathways, including hydrogen oxidation. J Bacteriol. 1989 Feb;171(2):1093–1099. doi: 10.1128/jb.171.2.1093-1099.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SILVERMAN M. P., LUNDGREN D. G. Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. I. An improved medium and a harvesting procedure for securing high cell yields. J Bacteriol. 1959 May;77(5):642–647. doi: 10.1128/jb.77.5.642-647.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196(3):413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. R., Cunningham R. P., Holmes D. S. IST2: an insertion sequence from Thiobacillus ferrooxidans. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7284–7287. doi: 10.1073/pnas.85.19.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]