Abstract
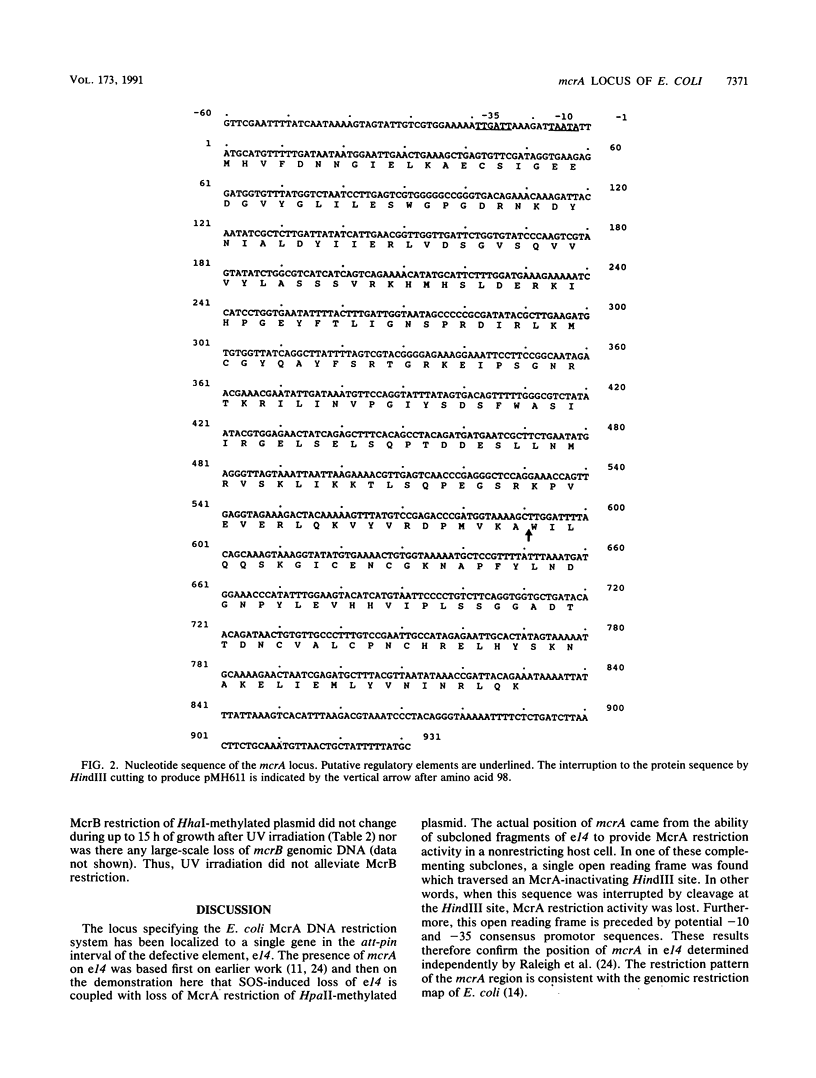
Escherichia coli has DNA restriction systems which are able to recognize and attack modified cytosine residues in the DNA of incoming bacteriophages and plasmids. The locus for the McrA/RglA system of modified cytosine restriction was located near the pin gene of the defective element, e14. Hence, loss of the e14 element through abortive induction after UV irradiation caused a permanent loss of McrA restriction activity. e14 DNA encoding McrA restriction was cloned and sequenced to reveal a single open reading frame of 831 bp with a predicted gene product of 31 kDa. Clones expressing the complete open reading frame conferred both McrA and RglA phenotypes; however, a deletion derivative was found which complemented RglA restriction against nonglucosylated T6gt phage but did not complement for McrA restriction of methylated plasmid DNA. Possible explanations for this activity and a comparison with the different organization of the McrB/RglB restriction system are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amasino R. M. Acceleration of nucleic acid hybridization rate by polyethylene glycol. Anal Biochem. 1986 Feb 1;152(2):304–307. doi: 10.1016/0003-2697(86)90413-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brody H., Hill C. W. Attachment site of the genetic element e14. J Bacteriol. 1988 May;170(5):2040–2044. doi: 10.1128/jb.170.5.2040-2044.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R. S., 3rd UV-induced alleviation of K-specific restriction of bacteriophage lambda. J Virol. 1977 Mar;21(3):1249–1251. doi: 10.1128/jvi.21.3.1249-1251.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dila D., Raleigh E. A. Genetic dissection of the methylcytosine-specific restriction system mcrB of Escherichia coli K-12. Gene. 1988 Dec 25;74(1):23–24. doi: 10.1016/0378-1119(88)90241-7. [DOI] [PubMed] [Google Scholar]
- Dila D., Sutherland E., Moran L., Slatko B., Raleigh E. A. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J Bacteriol. 1990 Sep;172(9):4888–4900. doi: 10.1128/jb.172.9.4888-4900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener A., Hill C. W. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980 Oct;144(1):312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K., Thomas S. M., Sedgwick S. G. Different mechanisms for SOS induced alleviation of DNA restriction in Escherichia coli. Biochimie. 1991 Apr;73(4):399–405. doi: 10.1016/0300-9084(91)90106-b. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- LURIA S. E., HUMAN M. L. A nonhereditary, host-induced variation of bacterial viruses. J Bacteriol. 1952 Oct;64(4):557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyer-Weidner M., Diaz R., Reiners L. Cytosine-specific DNA modification interferes with plasmid establishment in Escherichia coli K12: involvement of rglB. Mol Gen Genet. 1986 Dec;205(3):469–475. doi: 10.1007/BF00338084. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A., Yuan R., Stein D. C. Construction of a temperature-sensitive mutation for the direct identification of plasmids encoding DNA methyltransferases. Gene. 1988 Dec 25;74(1):233–235. doi: 10.1016/0378-1119(88)90294-6. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A., Yuan R., Stein D. C. Isolation of temperature-sensitive McrA and McrB mutations and complementation analysis of the McrBC region of Escherichia coli K-12. J Bacteriol. 1991 Jan;173(1):150–155. doi: 10.1128/jb.173.1.150-155.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REVEL H. R., HATTMAN S., LURIA S. E. MUTANTS OF BACTERIOPHAGES T2 AND T6 DEFECTIVE IN ALPHA-GLUCOSYL TRANSFERASE. Biochem Biophys Res Commun. 1965 Feb 17;18:545–550. doi: 10.1016/0006-291x(65)90788-6. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Benner J., Bloom F., Braymer H. D., DeCruz E., Dharmalingam K., Heitman J., Noyer Weidner M., Piekarowicz A., Kretz P. L. Nomenclature relating to restriction of modified DNA in Escherichia coli. J Bacteriol. 1991 Apr;173(8):2707–2709. doi: 10.1128/jb.173.8.2707-2709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A. Restriction and modification in vivo by Escherichia coli K12. Methods Enzymol. 1987;152:130–141. doi: 10.1016/0076-6879(87)52015-8. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Trimarchi R., Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989 Jun;122(2):279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R. S., Sozhamannan S., Dharmalingam K. Transposon mutagenesis and genetic mapping of the rglA and rglB loci of Escherichia coli. Mol Gen Genet. 1985;198(3):390–392. doi: 10.1007/BF00332928. [DOI] [PubMed] [Google Scholar]
- Revel H. R. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967 Apr;31(4):688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- Ross T. K., Achberger E. C., Braymer H. D. Characterization of the Escherichia coli modified cytosine restriction (mcrB) gene. Gene. 1987;61(3):277–289. doi: 10.1016/0378-1119(87)90191-0. [DOI] [PubMed] [Google Scholar]
- Ross T. K., Achberger E. C., Braymer H. D. Identification of a second polypeptide required for McrB restriction of 5-methylcytosine-containing DNA in Escherichia coli K12. Mol Gen Genet. 1989 Apr;216(2-3):402–407. doi: 10.1007/BF00334382. [DOI] [PubMed] [Google Scholar]
- Ross T. K., Achberger E. C., Braymer H. D. Nucleotide sequence of the McrB region of Escherichia coli K-12 and evidence for two independent translational initiation sites at the mcrB locus. J Bacteriol. 1989 Apr;171(4):1974–1981. doi: 10.1128/jb.171.4.1974-1981.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross T. K., Braymer H. D. Localization of a genetic region involved in McrB restriction by Escherichia coli K-12. J Bacteriol. 1987 Apr;169(4):1757–1759. doi: 10.1128/jb.169.4.1757-1759.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub F., Thompson E. B. An improved method for preparing large arrays of bacterial colonies containing plasmids for hybridization: in situ purification and stable binding of DNA on paper filters. Anal Biochem. 1982 Oct;126(1):222–230. doi: 10.1016/0003-2697(82)90133-6. [DOI] [PubMed] [Google Scholar]
- Thoms B., Wackernagel W. UV-induced allevation of lambda restriction in Escherichia coli K-12: kinetics of induction and specificity of this SOS function. Mol Gen Genet. 1982;186(1):111–117. doi: 10.1007/BF00422921. [DOI] [PubMed] [Google Scholar]
- Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990 Mar;54(1):18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Plasterk R., Kuijpers A. A Mu gin complementing function and an invertible DNA region in Escherichia coli K-12 are situated on the genetic element e14. J Bacteriol. 1984 May;158(2):517–522. doi: 10.1128/jb.158.2.517-522.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]