Abstract
A general approach for crystallization of proteins in a fast and simple manner would be of immense interest to biologists studying protein structure–function relationships. Here, we describe a method that we have developed for promoting the formation of helical arrays of proteins and macromolecular assemblies. Electron micrographs of the arrays are suitable for helical image analysis and three-dimensional reconstruction. We show that hydrated mixtures of the glycolipid galactosylceramide (GalCer) and derivatized lipids or charged lipids form unilamellar nanotubules. The tubules bind proteins in a specific manner via high affinity ligands on the polar head groups of the lipid or via electrostatic interactions. By doping the GalCer with a novel nickel-containing lipid, we have been able to form helical arrays of two histidine-tagged proteins. Similarly, doping with a biotinylated lipid allows crystallization of streptavidin. Finally, three proteins with affinity for positively or negatively charged lipid layers formed helical arrays on appropriately charged tubules. The generality of this method may allow a wide variety of proteins to be crystallized on lipid nanotubes under physiological conditions.
Keywords: tubules/helical arrays/galactosylceramides/electron microscopy
Electron microscopy (EM) has become an increasingly powerful method for three-dimensional (3D) structure determination of both relatively small and very large molecules and macromolecular assemblies. Advances in cryo-imaging (1, 2) along with faster and more sophisticated computer analysis of electron micrographs have allowed important structural information to be obtained from images of single particles, two-dimensional (2D) crystals, and helical arrays at high (3–10 Å) and moderate resolutions (10–40 Å). Whereas high resolution 3D maps may be interpreted directly in terms of the atomic structure, maps of macromolecular complexes at moderate resolution may be combined with x-ray structures of the individual components to yield near-atomic models of the entire complex. This combination of cryo-EM and x-ray crystallography has answered questions that could not be addressed by either technique alone (see, e.g., refs. 3 and 4).
So far, single particle analysis has been limited to large macromolecular assemblies. Icosahedral viruses, which have a high degree of symmetry, have been particularly suitable objects for study, and secondary structure elements have been visualized recently in hepatitis B virus cores by cryo-EM and image analysis (5, 6). Single particles that have low or no internal symmetry have yielded 3D maps in the 15–30 Å resolution range and have provided important insights into structure–function relationships in, for example, the ryanodine receptor/calcium channel and the ribosome (7–10).
Analysis of images of 2D crystals (11) has provided the most detailed information so far, and near-atomic resolution structures of several important biological molecules have been determined uniquely by this method. Examples include bacteriorhodopsin (11, 12), the plant light-harvesting complex (13), porin (14), and tubulin (15). 2D crystals are very useful because they generally contain a very large number of unit cells, and electron diffraction as well as imaging can be used to obtain high quality data sets. However, obtaining a high resolution structure by tilt series reconstruction and electron diffraction is a very time-consuming task, and the limits on tilting in the microscope cause the resolution in the final 3D map perpendicular to the plane of the crystal to be significantly poorer than the resolution in the plane of the crystal.
3D maps from helical specimens do not suffer from these limitations; tilting is not necessary because all required molecular views are present in a single image, and semi-automatic helical analysis and averaging allow the rapid calculation of 3D maps with isotropic resolution and high signal-to-noise ratios (16, 17). So far, no general method for helical crystallization has been available, and helical analysis has been limited to protein polymers that naturally adopt this organization or to proteins in which fortuitous helical crystallization has taken place. Although many helical specimens (e.g., actomyosin or microtubules) are ordered to only 20- to 30-Å resolution, important insights into function have been obtained from 3D maps at this resolution. Well ordered helical specimens such as tobacco mosaic virus (18), bacterial flagellar filaments (19), and acetylcholine receptor tubes (20) allow visualization of secondary structure elements in the 7- to 11-Å resolution range. With current developments in imaging and analysis, near-atomic resolution data seem within reach.
Sample preparation still remains the critical step for obtaining high resolution structural information of biological macromolecules. The methodology for preparing 2D crystals is maturing rapidly both for membrane proteins and for soluble proteins that have been ordered on lipid layers (reviewed in refs. 21 and 22). The latter method, introduced by Uzgiris and Kornberg (23), is particularly versatile because of the possibility of modifying the lipid substrate to facilitate protein binding. Streptavidin crystals grown on lipid layers by this method diffract to <3-Å resolution (24, 25).
In the work described here, we have combined the principles of the lipid layer crystallization methodology with the propensity of certain lipids to form nanotubes (26) to develop a general approach for helical crystallization of macromolecules. By incorporating charged lipids or lipids that have been chemically modified with high affinity ligands into the nanotubes, molecules from solution can be adsorbed specifically to the nanotube surface where crystallization is favored. By using this method, we have grown helical arrays of a variety of proteins. The arrays are suitable for helical analysis and may be used to derive structural information at moderate (≈20 Å) resolution. The major difficulty in EM structure determinations has been in preparing well ordered samples, so this methodology, incorporating the ease and speed of 3D helical analysis, offers an exciting alternative to existing lipid layer crystallization. Further development of this methodology may allow rapid and easy structure determinations at near atomic resolution.
METHODS AND MATERIALS
Lipids.
Initial studies were done by using a galactosylceramide containing a nervonyl (24:1Δ15(cis)) acyl chain (26). Mixed galactosylceramides and sphingosine were purchased from Sigma. The nickel lipid DOGS-NTA-Ni was custom synthesized by Avanti Polar Lipids. The structure was confirmed by MS and was 99% pure by TLC and GC (assays carried out by Avanti Polar Lipids). Charged lipids DO-ETHYL-PC, DOTAP, and DOPS were purchased from Avanti Polar Lipids. Biotinylated lipid, Biotin-X-DPPE, was purchased from Molecular Probes.
Proteins.
Annexin V was purchased from Sigma. Streptavidin was purchased from Boehringer Mannheim. Escherichia coli RNA polymerase was kindly provided by Seth Darst (Rockefeller). His-tagged Fab AP7 (27) was provided by Tom Kunicki (Scripps). His-tagged Fab 3B3 (28) was provided by Erica Ollmann and Ian Wilson (Scripps). Actin was prepared from rabbit skeletal muscle (29) and was provided by Mike Whittaker (Scripps).
Tubule Preparation.
Aliquots (10–30 μl) of synthetic GalCer in chloroform/isopropanol/water (70:30:1) at 5–10 mg/ml were dried down under argon and rehydrated in buffer at ≈1 mg/ml. Nickel–lipid-containing and charged tubules were prepared in a similar manner by mixing 10–80% DOGS-NTA-Ni or 10–30% charged lipids in chloroform with either synthetic GalCer or crude GalCer (Sigma) in chloroform/methanol (1:1). The solutions then were dried down and resuspended in buffer at ≈1 mg/ml. The above lipid mixtures were sonicated for 1–6 min in a water bath at room temperature.
Helical Arrays on Lipid Tubules.
Helical arrays were formed by incubating his-tagged Fab AP7 (≈50 μg/ml) with GalCer tubules doped with 20% DOGS-NTA-Ni (≈0.5 mg/ml) in 20 mM Hepes (pH 7.0) and 100 mM NaCl. Conditions for helical crystallization of his-tagged Fab 3B3 were similar except that the tubules contained 10% DOGS-NTA-Ni (0.8 mg/ml). Streptavidin (≈25 μg/ml) formed helical arrays when incubated with tubules containing 10% biotin-X-DPPE (0.8 mg/ml) in 50 mM Tris⋅HCl (pH 7.5) and 200 mM NaCl. E. coli RNA polymerase (≈100 μg/ml) formed helical arrays when incubated with tubules containing 10% DO-Ethyl-PC (0.2 mg/ml) in 50 mM Tris·HCl (pH 7.8) and 100 mM ammonium acetate. Experiments with f-actin and phalloidin-stabilized f-actin were carried out at a protein concentration of ≈200 μg/ml in 10 mM Hepes (pH 7.0), 50 mM KCl, and 2 mM MgCl2 by using tubules containing 10% DO-Ethyl-PC (1 mg/ml). G-actin at 100 μg/ml in 50 mM Tris·HCl (pH 7.8) and 100 mM ammonium acetate formed helical arrays on the same tubules. Annexin V at ≈100 μg/ml in 20 mM Tris·HCl (pH 7.0), 100 mM NaCl, and 1 mM CaCl2 formed helical arrays on tubules containing 10–30% DOPS (1 mg/ml).
EM and Image Analysis.
After incubation, 5-μl aliquots of the solutions were applied to glow-discharged support films on 400 mesh copper EM grids. Grids either were negatively stained with 1% uranyl acetate or were blotted and frozen for cryo-EM. Electron micrographs were recorded under low dose conditions (<10 e/Å2) with the exception of the electron micrograph of the helical array of Fab AP7, which was a high dose image. Philips CM100, CM120, or CM200 electron microscopes operating at 100 kV were used to record images at nominal magnifications of 35,000 or 38,000. Suitable images were digitized on a flat bed microdensitometer (Perkin–Elmer) with spot and step sizes of 20 μm (5.26 or 5.71 Å at the specimen). Some tubule images were straightened computationally, and computed diffraction patterns were calculated from well ordered areas by using modules from the program package phoelix (17).
RESULTS
Initially we used a synthetic chain-pure galactosylceramide (GalCer) with a nervonyl (24:1Δ15(cis)) acyl chain moiety that was reported to form nanotubes in aqueous solutions (26). During the course of our studies, we also used a commercially available bovine brain GalCer mixture that was less pure to achieve similar results. We refer to tubules that were prepared with either galactosylceramide preparation as “GalCer tubules.” The tubules, preserved in negative stain or vitreous ice, were observed by EM. Tubules prepared with GalCer alone tended to clump together and form a precipitate (data not shown). By adding specific lipids [e.g., nickel-lipid, DOGS-NTA-Ni (Fig. 1a), a biotinylated lipid, biotin-X-DPPE), or charged lipids (e.g., positively charged DO-Ethyl-PC, negatively charged, DOPS)] to the GalCer, the tubules were less aggregated and we could readily form lipid nanotubes in a physiological buffer (see Materials and Methods).
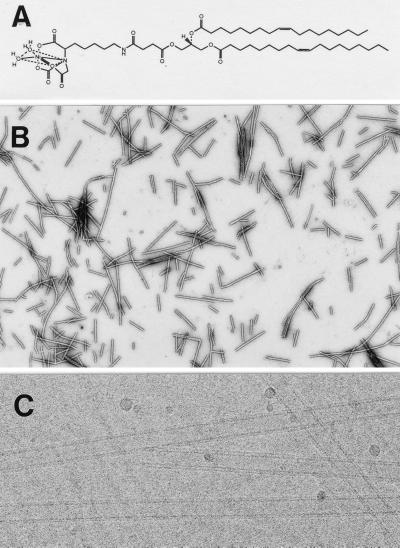
Figure 1.
Characterization of lipid tubules. (A) Chemical structure of the nickel lipid (DOGS-NTA-Ni) used in this study. (B) Nickel-functionalized lipid tubules negatively stained with 1% uranyl acetate: 90% GalCer, 10% DOGS-NTA-Ni. (Magnification ×7,600.) (C) Unstained, negatively charged lipid tubules preserved in vitrified buffer: 90% GalCer, 10% DOPS. The tubules are unilamellar with an inner diameter ≈160 Å and an outer diameter of ≈270 Å. (Magnification ×140,000.)
Low magnification images of a mixture containing 90% GalCer and 10% DOGS-NTA-Ni (wt/wt) preserved in negative stain revealed mainly tubular structures up to several microns long (Fig. 1b). A high magnification image of the nanotubules preserved in vitreous ice showed that they are unilamellar and of fairly uniform diameter (≈270 Å) (Fig. 1c). Surprising to note, only relatively low concentrations of GalCer were, in some cases, required for tubule formation; solutions containing 80% nickel lipid and 20% GalCer form tubules. With <20% GalCer, the solution contained mostly vesicles and sheets. No tubules formed with the nickel lipid alone. Tubules incorporating charged lipids were less stable, and in some cases incorporation of >10% charged lipid prevented tubule formation. Tubules were consistently longer when 5% polyethylene glycol 2,000 was added to the buffer solution. Tubule formation did not appear to be affected by concentrations of NaCl up to 500 mM. However, when divalent cations such as Mg2+ were present at >10 mM, no tubules were observed. We have not studied other lipid structures that were observed by EM but concentrated our efforts on exploring the utility of the doped tubules.
Nickel–Histidine-Mediated Attachment and Helical Crystallization.
The polar head groups of lipids may be modified by covalent attachment of high affinity ligands to allow specific binding of a receptor that has been incorporated into recombinant proteins. Nickel lipids provide a common moiety that allows specific binding of histidine tagged (his-tagged) proteins. This interaction has been used to adsorb protein directly to planar lipid surfaces facilitating the formation of 2D crystals (30–32). We used a nickel lipid (DOGS-NTA-Ni, Fig. 1a) to functionalize the GalCer tubules. Tubules containing 10% DOGS-NTA-Ni were used to crystallize two his-tagged Fab fragments (≈50 μg/ml protein in 20-μl aliquots). Both antibodies bound to nickel–lipid-doped tubules. Neither bound to 100% GalCer tubules nor to doped tubules in the presence of 100 mM imidazole, demonstrating that binding is specifically due to the histidine–nickel interactions (results not shown).
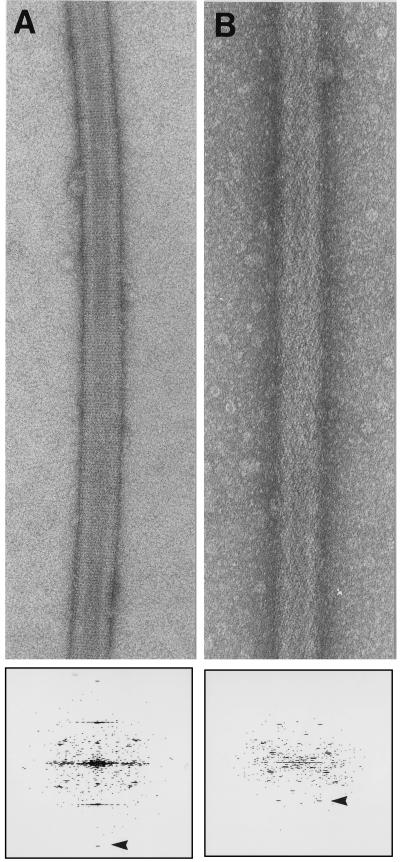
We chose antibody fragments that were tagged at the N terminus or the C terminus of the heavy chain to explore what effect the tag position might have on the crystalline lattice. Fab 3B3 has the his-tag on the N terminus whereas Fab AP7 has the tag on the C terminus. Of interest, both the images and the diffraction patterns show that the antibodies did not crystallize in the same way, instead forming quite different helical packing arrangements on the surface of the tubules, (Fig. 2 a and b). This result suggests that his-tags in different locations may be used to localize regions of the protein or to facilitate crystallization (30).
Figure 2.
Helical arrays of two his-tagged proteins on nickel functionalized lipid tubules. (A) Helical array of his tagged Fab 3B3 grown on a nickel functionalized lipid tubule, negatively stained with 1% uranyl acetate. The diffraction pattern below shows visible peaks to 1/19 Å−1 (arrowhead). (Magnification ×140,000.) (B) Helical array of his tagged Fab AP7 grown on a nickel-functionalized lipid tubule, negatively stained with 1% uranyl acetate. The diffraction pattern below shows visible peaks to 1/30 Å−1 (arrowhead). (Magnification ×152,000.)
Streptavidin-Biotin Mediated Attachment and Helical Crystalization.
It is also possible to take advantage of natural high affinity sites in a protein, as in the case of streptavidin, which has four biotin binding sites. When streptavidin interacts with a biotinylated lipid layer, two of its binding sites are occupied, while the remaining sites are left unoccupied, available to bind biotin moieties from solution. A streptavidin crystal formed on a lipid tubule thus could serve as a scaffold for further attachment of proteins. A mixture of biotin-X-DPPE with GalCer produced tubules that readily bound streptavidin (≈25 μg/ml protein in 20-μl aliquots) and formed highly ordered helical arrays (Fig. 3). Similar results have been obtained by Brisson and colleagues by using a biotin lipid (DODA-EO2-biotin) that forms tubules without GalCer (33).
Figure 3.
Unstained GalCer tubule containing 10% biotinylated lipid showing helical array of streptavidin, preserved in vitrified buffer. The diffraction pattern below shows visible peaks to 1/27 Å−1 (arrowhead). (Magnification ×152,000.)
Protein Binding and Helical Crystallization on Positively Charged Tubules.
More generally, tubules can be formed that use relatively nonspecific electrostatic interactions between charged lipid head groups and accessible charged residues on proteins to facilitate helical crystallization. It has been demonstrated that proteins that have surface charges have formed 2D crystals on lipids with the opposing charge. We tested two proteins, E. coli RNA polymerase (34, 35) and f-actin (36, 37), that form ordered arrays on positively charged lipid layers for their ability to form helical arrays on the tubules.
Although we tested a number of positively charged lipids, only DO-Ethyl-PC was successful in supporting the formation of ordered arrays of both proteins. Some of the positively charged lipids we tested allowed protein binding but were not successful in forming helical arrays; others produced difficulties in tubule formation. Under similar ionic conditions as described by Darst and colleagues (38), it was possible to form helical arrays of RNA polymerase on positively charged DO-Ethyl-PC tubules (≈100 μg/ml protein in aliquots of 20 μl) (Fig. 4a). RNA polymerase bound to GalCer tubules doped with either 10% DOTAP or 10% sphingosine, but no ordering was observed. Control experiments show that negatively charged tubules made by doping GalCer tubules with 10% DOPS did not bind polymerase (data not shown). Thus, polymerase binding depends on the presence of a positively charged lipid. Previously, it was shown that polymerase will not bind to neutral or negatively charged lipid tubules (34).
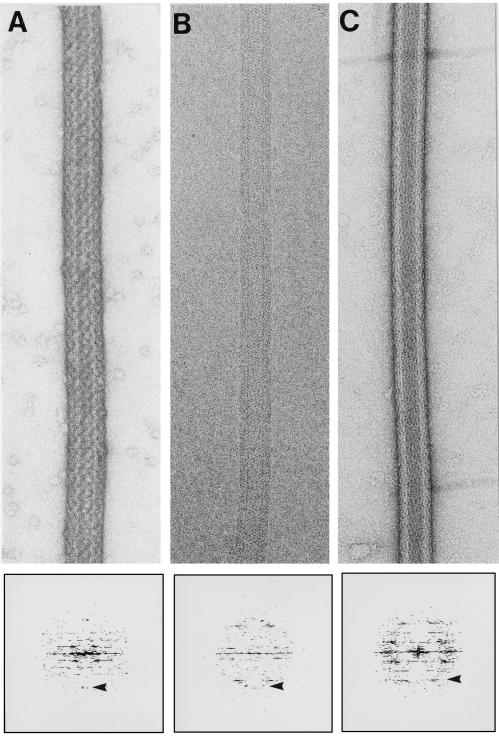
Figure 4.
Helical arrays of a macromolecular complex and two proteins formed on charged lipid tubules. (A) Helical array of RNA polymerase on a DO-Ethyl-PC functionalized lipid tubule (positive surface charge), negatively stained with 1% uranyl acetate. The diffraction pattern below shows visible peaks to 1/38 Å−1 (arrowhead). (Magnification ×140,000.) (B) Cryo-electron micrograph of a helical array of g-actin on DO-Ethyl-PC functionalized lipid tubule (positive surface charge). The diffraction pattern below shows visible peaks up to 1/35 Å−1 (arrowhead). (Magnification ×152,000.) (C) Helical array of annexin V on DOPS functionalized lipid tubule (negative surface charge) negatively stained with 1% uranyl acetate. The diffraction pattern below shows visible peaks up to 1/40 Å−1 (arrowhead). (Magnification ×152,000.)
When a solution of rabbit skeletal muscle f-actin was incubated with tubules doped with 10% DO-Ethyl-PC a variety of poorly ordered arrays were formed (data not shown). The addition of phalloidin, which stabilizes filaments and lowers the critical concentration for polymerization, to the f-actin solution abolished formation of these arrays and the actin filaments bound to the tubules in a parallel fashion without apparent ordering (data not shown). However, when unpolymerized actin (g-actin) was used (≈100 μg/ml protein in aliquots of 20 μl), extensive ordering was observed on the surface of the positively charged tubules (Fig. 4b). Taken together, these results suggest that g-actin, but not f-actin, is capable of forming arrays on the tubules. The helical arrays on the tubules are therefore distinct from the paracrystalline sheets of f-actin formed on planar lipid films (36, 37).
Protein Binding and Helical Crystallization on Negatively Charged Tubules.
Finally, we examined the ability of negatively charged tubules to facilitate binding and crystallization of annexin V. This protein forms 2D crystals when incubated with negatively charged planar lipid layers (39). We incubated annexin V (≈100 μg/ml protein in aliquots of 20 μl) with tubules made from 90–70% GalCer and 10–30% DOPS under ionic conditions similar to those used by Brisson and colleagues to grow 2D lipid layer crystals (39). Extensive helical ordering was observed (Fig. 4c).
DISCUSSION
We have produced specifically and nonspecifically functionalized unilamellar lipid tubules by using mixtures of a tubule-forming lipid (GalCer) and various derivatized or charged lipids. The functionalized tubules promoted binding and helical crystallization of several test proteins. When the polar head group of the doping lipid is derivatized with a high affinity ligand, specific binding via the corresponding receptor or tag becomes possible and promotes the formation of ordered arrays. High affinity couples such as poly histidine–nickel are used commonly to facilitate protein purification from expression systems, and we successfully have adapted this interaction for our helical crystallization system. We have shown that two his-tagged Fab fragments form helical arrays on the surface of GalCer tubules containing the nickel lipid DOGS-NTA-Ni. This nickel lipid is readily incorporated into lipid tubules when mixed with GalCer. One of the Fabs used has the his-tag at the C terminus of the heavy chain. The other Fab has the his-tag at the N terminus. Presumably, the different helical lattices seen with the two Fabs are a consequence of the different orientations of the protein on the tubule surface, which may allow one to localize in 3D maps the region of the protein that is attached to the surface (30). A number of variations on this interaction scheme are possible. For example, a copper chelating lipid has been used to mediate formation of 2D crystals of a protein on lipid layers via surface accessible histidines, demonstrating that it may not be necessary to use recombinant methods to introduce poly histidine tags (42). This variation easily could be adapted for our helical crystallization approach.
We also show that one can take advantage of natural high affinity sites in a protein as in the case of streptavidin, which has four biotin binding sites. The biotinylated lipid we used and the DODA-EO2 biotin lipid used by Brisson and colleagues (33) produced tubules that promoted the formation of helical arrays of streptavidin. The two biotin sites of the streptavidin molecule that are not bound to the lipid and are presumably on the outside of the tubular protein array potentially could be used to bind other biotinylated proteins. In this way, the existing streptavidin helical array might be used as a substrate for further crystallization of biotinylated molecules (33, 40). Disadvantages of this approach for crystallization are that the second layer of proteins has to comply with the underlying streptavidin lattice and that the molecules may not have the freedom of rotation that is needed to crystallize. Although this particular approach has not yet met with success, a similar concept has been explored by Ren et al. (41). They used the high affinity of T4 small outer capsid protein for gp23* in capsids and polyheads to mediate small outer capsid–fusion protein attachment to the capsid/polyhead lattice. The small outer capsid–fusion protein was ordered on the underlying gp23* lattice. It was concluded that the substrate lattice could accommodate proteins as large as 35 kDa. These results suggest that it may be possible to develop a general approach for substrate–lattice crystallization.
We have shown that it is also possible to take advantage of the intrinsic properties of proteins (e.g., surface charge) to promote binding and the formation of helical arrays on tubules. We show that relatively small proteins such as actin (43 kDa) and annexin V (35 kDa), as well as a large macromolecular complex, RNA polymerase (450 kDa), form helical arrays on charged unilamellar lipid tubules. Crystalline sheets of monomeric actin can be formed in solution by the addition of trivalent lanthanides (43), but we demonstrate that g-actin can crystallize on a lipid surface. F-actin also bound to the tubules but, under the conditions we used, did not form an ordered array. RNA polymerase bound to a variety of positively charged lipid tubules, but helical arrays were found with only one of the positively charged lipids tested. This type of observation was not unique to charged lipid tubules; N-ethylmaleimide-sensitive factor, which has been shown to bind to Ni-NTA-DOPE lipid (44), bound specifically to nickel–lipid-containing tubules but did not form ordered arrays. So, although concentrating the protein at the interfacial adsorption surface is facilitated by either electrostatic or specific high affinity interactions, the results of this study demonstrate that the lipid head group also appears to play a critical role in the way the protein molecules bind and align themselves into ordered arrays. Future experiments will explore the effects of varying the spacing between the nickel chelating head group and the glycerol backbone on crystallization of bound molecules.
An advantage of the approach described here is that it allows one to do many crystallization trials very rapidly. Each experiment requires only microgram quantities of protein. The lipid tubules are stable in a variety of buffers and can be produced simply within minutes. Crystallization takes place in an accessible solution, so it is possible to vary the conditions at any point during the experiment. The helical arrays formed on the lipid tubules are ideal specimens for 3D electron microscopy. A drop containing the tubules can be placed directly on an EM support film and preserved in either negative stain or vitreous ice for examination and imaging. There are no transfer problems such as those described for 2D crystals formed by lipid layer crystallization (24). The arrays on the tubules are helical, so rapid calculation of high quality 3D maps is possible without tilting the specimen. Furthermore, the resulting 3D maps will not be distorted by the effects of anisotropic resolution, a problem encountered when analyzing 2D crystals. The tubules appear to be fairly uniform in width, suggesting that the surface arrays probably belong to a very limited set of helical families and making it easier to collect large data sets for averaging.
Here, we have shown a representative diffraction pattern for each of the helical protein arrays. The patterns show order in the 20- to 40-Å resolution range. We emphasize that we have not optimized conditions for growing any of the arrays. Neither have we used averaging and helical reconstruction methods because the object of this work was to develop a method for rapid and simple helical crystallization that would be applicable to a large variety of proteins. It therefore seems likely that the order of many, if not all, of the helical arrays could be improved dramatically by optimizing the solution conditions and by computational correction of the images. The data presented here establish proof of concept for the methodology and lay the foundation for developing a diverse bank of functionalized unilamellar lipid tubules that are stable over a wide range of conditions. Such a bank of tubules would allow rapid screening of virtually any protein or macromolecular complex for helical crystallization.
Lipid tubules have been studied for over a decade because of their potential applications in nanotechnology. The nanotubules we have produced may serve as a tool, either coated with proteins or uncoated, for in vivo and in vitro studies using a helical nucleation surface. Biomineralization is one area with interesting possibilities. Nucleation of inorganic crystals on the external surface of lipid microstructures has been reported, but unilamellar tubules could not be produced (45). The tubules we have used are predominantly unilamellar, have a relatively constant diameter, and can be prepared readily with either a positive or negative surface charge. It therefore may be possible to mineralize them by nucleation and coating with a mineral such as calcite. A new range of inorganic–organic materials can be envisioned.
Acknowledgments
We are grateful to Ian Wilson and Erica Ollmann (Scripps) for providing Fab 3B3, Tom Kunicki (Scripps) for Fab AP7, Mike Whittaker (Scripps) for actin, and Seth Darst (Rockefeller) for E. coli RNA polymerase. We thank Mike Whittaker for help with computation and Gavin Meredith for critical comments on the manuscript. This work was supported in part by grants from the National Institutes of Health (AR39155, AR44278, and GM52468 to R.A.M. and GM45928 to R.E.B.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: 3D, three-dimensional; 2D, two-dimensional; GalCer, galactosylceramide; DO-Ethyl-PC, 1,2-dioleoyl-sn-glycero-3-ethylphosphocoline; DOPS, 1,2-dioleoyl-sn-glycero-3-[phospho-l-serine]; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; DOGS-NTA-Ni, 1,2-dioleoyl-sn-glycero-3-[(N(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] (Nickel salt); biotin-X-DPPE, N((6(biotinoyl)amino)hexanoyl) dipalmitoyl-l-α-phosphatidylethanolamine; his-tagged, poly histidine-tagged; EM, electron microscopy.
A commentary on this article begins on page 7848.
References
- 1.Taylor K, Glaeser R M. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 2.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall A W, Schultz P. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 3.Rayment I, Holden H, Whittaker M, Yohn C, Lorenz M, Holmes K, Milligan R. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 4.Sosa H, Dias D P, Hoenger A, Whittaker M, Wilson-Kubalek E M, Sablin E, Fletterick R J, Vale R D, Milligan R A. Cell. 1997;90:217–224. doi: 10.1016/s0092-8674(00)80330-x. [DOI] [PubMed] [Google Scholar]
- 5.Bottcher B, Wynne S A, Crowther R A. Nature (London) 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 6.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Nature (London) 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 7.Rademacher M, Rao V, Grassucci R, Frank J, Timmerman A P, Fleischer S, Wagenknecht J Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlova E V, Serysheva I, I, van Heel M, Hamilton S L, Chiu W. Nat Struct Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- 9.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Vershoor A, Radimacher M, Grassucci R, Lata K R, Agrawal R K. Nature (London) 1995a;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 10.Stark H, Mueller F, Orlova E V, Schaltz M, Dube P, Erdermir T, Zemlin F, Brimacombe R, van Heel M. Structure. 1995;3:815–821. doi: 10.1016/s0969-2126(01)00216-7. [DOI] [PubMed] [Google Scholar]
- 11.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 12.Ceska T A, Henderson R, Baldwin J M, Zemlin F, Beckmann E, Downing K H. Acta Physiol Scand Suppl. 1992;607:31–40. [PubMed] [Google Scholar]
- 13.Kuehlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 14.Jap B K, Walian P J, Gehring K. Nature (London) 1991;350:167–170. doi: 10.1038/350167a0. [DOI] [PubMed] [Google Scholar]
- 15.Nogales E, Wolf S G, Downing K H. Nature (London) 1998;391:199–202. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 16.Morgan D G, DeRosier D. Ultramicroscopy. 1992;46:263–285. doi: 10.1016/0304-3991(92)90019-g. [DOI] [PubMed] [Google Scholar]
- 17.Whittaker M, Carragher B O, Milligan R A. Ultramicroscopy. 1995;58:245–260. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]
- 18.Jeng T-W, Crowther R A, Stubbs G, Chiu W. J Mol Biol. 1989;205:251–257. doi: 10.1016/0022-2836(89)90379-3. [DOI] [PubMed] [Google Scholar]
- 19.Morgan D G, Owen C, Melanson L A, DeRosier D. J Mol Biol. 1995;249:88–110. doi: 10.1006/jmbi.1995.0282. [DOI] [PubMed] [Google Scholar]
- 20.Unwin N. Nature (London) 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- 21.Jap B K, Zulauf M, Scheybani T, Hefti A, Baumeister W, Aebi U, Engel A. Ultramicroscopy. 1992;46:45–84. doi: 10.1016/0304-3991(92)90007-7. [DOI] [PubMed] [Google Scholar]
- 22.Chiu W, Avila-Sakar A J, Schmid M F. Adv Biophys. 1997;34:161–172. doi: 10.1016/s0065-227x(97)89638-4. [DOI] [PubMed] [Google Scholar]
- 23.Uzgiris E E, Kornberg R D. Nature (London) 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 24.Kubalek E W, Kornberg R D, Darst S A. Ultramicroscopy. 1991;35:295–304. doi: 10.1016/0304-3991(91)90082-h. [DOI] [PubMed] [Google Scholar]
- 25.Avila-Sakar A J, Chiu W. Biophys J. 1996;70:57–68. doi: 10.1016/S0006-3495(96)79597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkarni V S, Anderson W H, Brown R E. Biophys J. 1995;69:1976–1986. doi: 10.1016/S0006-3495(95)80068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunicki T J, Annis D S, Felding-Habermann B. J Biol Chem. 1997;272:4103–4107. doi: 10.1074/jbc.272.7.4103. [DOI] [PubMed] [Google Scholar]
- 28.Barbas C F, 3rd, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry R M, Burton D R. Proc Natl Acad Sci USA. 1994;97:3809–3813. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spudich J A, Watt S. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 30.Kubalek E W, Le Grice S F J, Brown P O. J Struct Biol. 1994;113:117–123. doi: 10.1006/jsbi.1994.1039. [DOI] [PubMed] [Google Scholar]
- 31.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid M F, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, et al. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venien-Bryan C, Balavoine F, Toussaint B, Mioskowski C, Hewat E A, Helme B, Vignais P M. J Mol Biol. 1997;274:687–692. doi: 10.1006/jmbi.1997.1431. [DOI] [PubMed] [Google Scholar]
- 33.Ringler P, Muller W, Ringsdorf H, Brisson A. Chem Eur J. 1997;3:620–625. [Google Scholar]
- 34.Darst S A, Ribi H O, Pierce D W, Kornberg R D. J Mol Biol. 1988;203:269–273. doi: 10.1016/0022-2836(88)90107-6. [DOI] [PubMed] [Google Scholar]
- 35.Darst S A, Kubalek E W, Kornberg R D. Nature (London) 1989;340:730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- 36.Rioux L, Gicquaud C. J Ultrastruct Res. 1985;93:42–49. doi: 10.1016/0889-1605(85)90084-9. [DOI] [PubMed] [Google Scholar]
- 37.Taylor K A, Taylor D W. J Struct Biol. 1992;108:140–147. doi: 10.1016/1047-8477(92)90013-z. [DOI] [PubMed] [Google Scholar]
- 38.Polyakov A, Severinova E, Darst S A. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 39.Mosser G, Ravanat C, Freyssinet J-M, Brisson A. J Mol Biol. 1991;217:241–245. doi: 10.1016/0022-2836(91)90538-h. [DOI] [PubMed] [Google Scholar]
- 40.Darst S A, Ahlers M, Kubalek E W, Meller P, Blankenburg R, Ribi H O, Ringsdorf H, Kornberg R D. Biophys J. 1990;59:387–396. doi: 10.1016/S0006-3495(91)82232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren Z J, Lewis G K, Wingfield P T, Locke E G, Steven A C, Black L W. Protein Sci. 1996;5:1833–1843. doi: 10.1002/pro.5560050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey W, Schief W R, Jr, Pack D W, Chen C-T, Chilkoti A, Stayton P, Vogel V, Arnold F H. Proc Natl Acad Sci USA. 1996;93:4937–4941. doi: 10.1073/pnas.93.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aebi U, Fowler W E, Isenberg G, Pollard T D, Smith P R. J Cell Biol. 1981;91:340–351. doi: 10.1083/jcb.91.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteheart S W, Kubalek E W. Trends Cell Biol. 1995;5:64–68. doi: 10.1016/s0962-8924(00)88948-5. [DOI] [PubMed] [Google Scholar]
- 45.Archibald D D, Mann S. Nature (London) 1993;364:430–433. [Google Scholar]