Abstract
We have previously shown that the degradation of c-myc and N-myc in vitro is mediated by the ubiquitin system. However, the role of the system in targeting the myc proteins in vivo and the identity of the conjugating enzymes and possible ancillary proteins involved has remained obscure. Here we report that the degradation of the myc proteins in cells is inhibited by lactacystin and MG132, two inhibitors of the 20S proteasome. Inhibition is accompanied by accumulation of myc-ubiquitin conjugates. Dissection of the ancillary proteins involved revealed that the high-risk human papillomavirus oncoprotein E6-16 stimulates conjugation and subsequent degradation of the myc proteins in vitro. Expression of E6-16 in cells results in significant shortening of the t1/2 of the myc proteins with subsequent decrease in their cellular level. Analysis of the conjugating enzymes revealed that under basal conditions the proteins can be conjugated by two pairs of E2s and E3s—E2-14 kDa and E3α involved in the “N-end rule” pathway, and E2-F1 (UbcH7) and E3-Fos involved also in conjugation of c-Fos. In the presence of E6-16, a third pair, E2-F1 and E6-AP mediate conjugation of myc by means of a mechanism that appears to be similar to that involved in the targeting of p53, formation of a myc⋅E6⋅E6–AP targeting complex. It is possible that in certain cells E6-mediated targeting of myc prevents myc-induced apoptosis and thus ensures maintenance of viral infection.
The myc family of proteins is a group of structurally related transcriptional factors involved in a variety of cellular regulatory processes. They all contain basic helix–loop–helix (1) and leucine zipper (2) motifs. In many cases myc acts as a heterodimer with another protein, Max, which also contains basic helix–loop–helix and leucine zipper motifs (3). The c-myc oncogene has been implicated in control of normal cell proliferation. It is rapidly induced in resting cells following mitogenic stimulation, suggesting that it plays an important role in the transition from quiescence to proliferation. In tumor cells, elevated or deregulated expression of c-myc and N-myc has been observed frequently, suggesting a role for the proteins in malignant transformation (4). In many cases, the coexpression of myc with another oncoprotein, Ras, is important for its transforming ability. However, when cell proliferation is inhibited, deregulated c-myc expression can induce apoptosis. Thus, when grown on low serum, Rat-1 fibroblasts that express c-myc constitutively undergo rapid apoptosis (5). When the interleukin 3-dependent myeloid cells 32D, which also express c-myc constitutively, are deprived of the cytokine, they rapidly initiate a program of cell death (6). It is possible that apoptosis serves as an important mechanism in the elimination of cells harboring mutations, such as overexpression of c-myc, which under certain conditions imbalance cell cycle regulatory mechanisms.
The many functions that myc proteins play under different pathophysiological conditions imply that the cellular level of these proteins must be tightly regulated. Analysis of the c-myc gene revealed an important role for transcriptional regulation by means of its two major promoters, P1 and P2 (7). However, it has been shown that both the mRNA and the protein have extremely short half-lives, of 15 min (8) and 30 min (9), respectively. Thus, it appears that posttranscriptional regulatory mechanisms, including degradation, play important roles in regulating the level of myc proteins.
Recent evidence indicates that the ubiquitin proteolytic system plays a major role in targeting short-lived key regulatory proteins for degradation (10–12); among these are cyclins, tumor suppressors and oncoproteins, transcriptional activators, and endoplasmic reticulum and cell surface membrane proteins. Degradation of a protein by means of the ubiquitin system involves two discrete steps, conjugation of multiple molecules of ubiquitin to the target protein and degradation of the tagged substrate by the 26S proteasome. Conjugation proceeds in a three-step mechanism. Initially, ubiquitin is activated in its C-terminal Gly by the ubiquitin-activating enzyme (E1). Following activation, one of several ubiquitin-conjugating enzymes (E2s) transfers ubiquitin from E1 to a member of the ubiquitin-protein ligase (E3) family of enzymes to which the substrate protein is specifically bound. This enzyme catalyzes the last step in the conjugation process, covalent attachment of ubiquitin to the substrate and generation of a polyubiquitin chain anchored to an ɛ-NH2 group of a Lys residue of the protein substrate. The binding of the substrate to E3 is specific and implies that E3s play a major role in recognition and selection of proteins for conjugation. An important problem involves the mechanisms that underlie specific recognition of the many cellular substrates of the system. A few proteins may be recognized by means of their free N-terminal residue (“N-end rule”; ref. 13). However, most proteins are recognized by other signals. Some are targeted by sequences that reside downstream from the N-terminal residue. Others are targeted only following a specific posttranslational modification, such as phosphorylation, or following association with ancillary proteins, molecular chaperones for example (14). Recent information indicates that certain viral proteins can specifically target for degradation cellular proteins that may interfere with the ability of the virus to replicate. The human papillomavirus (HPV) high-risk oncoprotein E6-16 targets the tumor suppressor protein p53 for accelerated degradation by means of a specific ubiquitin–p53 ligase, E6-AP (15). Here, the viral protein prevents, most probably, p53-induced apoptosis and contributes, by means of the removal of a tumor suppressor, to the malignant transformation of the infected cells and to the continuity of viral replication and infection.
It should be also noted that recognition of protein substrates by the ubiquitin system does not always proceed by means of a “simple,” three-step E1–E2–E3 cascade. Several proteins are targeted by several specific pairs of E2 and E3 enzymes that appear to recognize distinct structural motifs. For example, the yeast transcriptional repressor MATα2 has two degradation signals, DEG1 and DEG2, and is recognized by four E2 enzymes, Ubc4, -5, -6, and -7 (16). Similarly, the model substrate lysozyme can be targeted by means of the “N-end rule” pathway by E2-14 kDa and E3α, but also, following recognition of a yet to be identified downstream motif, by E2-F1 and E3L (17).
MATERIALS AND METHODS
Chemicals.
Liposome-mediated transfection reagent (DOTAP) was from Boehringer Mannheim, l-[35S]methionine from New England Nuclear, ubiquitin from Sigma, and DEAE cellulose (DE52) from Whatman. All tissue culture reagents were purchased from Biological Industries (Bet Haemek, Israel). Geneticin (G418 sulfate) was from Life Technologies (Gaithersburg, MD). Antibodies against c-myc and N-myc were from Santa Cruz Biotechnology. Lactacystin and MG132 (N-carbobenzoxy-Leu-Leu-leucinal) were from Calbiochem. Restriction and modifying enzymes were from New England Biolabs. Immobilized protein A was from Pharmacia. All other reagents were of high analytical grade.
Plasmids.
cDNAs for in vitro translation of murine wild-type p53 and human N-myc were described elsewhere (18). cDNA encoding h-c-myc (18) for both in vitro translation (under the control of T7 RNA polymerase) and expression in mammalian cells was subcloned into the pCI-neo vector (Promega). cDNAs for in vitro translation of HPV E6-16 and E6-11 in pGEM-2 and pGEM-1 (Promega), for bacterial expression in pGEX-2T (Pharmacia) and for expression in mammalian cells in pRc/CMV (Invitrogen), were obtained from Martin Scheffner. Bacterial expression plasmid encoding the glutathione S-transferase (GST)-h-N-myc was constructed by subcloning of the N-myc cDNA into the pGEX-2TK vector (Pharmacia). Excision with NotI and EcoRI released the third exon of the molecule. Following filling in with Klenow enzyme and blunt-end ligation, we obtained a truncated N-myc protein that spans amino acid residues 1–232. Recombinant baculovirus and bacterial expression vector for E6-AP were obtained from Jon Huibregtse, Martin Scheffner, and Peter Howley.
Cell Lines.
The neuroblastoma cell line NGP, which expresses high levels of the human N-myc protein, was obtained from Garrett M. Brodeur.
In Vitro Translation.
cDNAs were transcribed/translated in wheat germ or reticulocyte extracts (TNT; Promega) in the presence of unlabeled or l-[35S]methionine according to the manufacturer’s instructions. The proteins were partially purified and the unreacted labeled amino acid was removed following chromatography over DEAE cellulose as described (18).
Preparation of Reticulocyte Extracts, Conjugating Enzymes, and E6 Proteins.
Reticulocytes were induced in rabbits and lysates were prepared and further fractionated into unadsorbed material (Fraction I) and high salt eluate (Fraction II) as described (19). Purified E1 and E2-14 kDa were prepared as described (19). E3α was prepared as described (20). E2-F1 was prepared as described (21). For characterization of the E3 enzymes, Fraction II was further fractionated by ammonium sulfate into Fraction IIA (0–38%), which contains all the known species E3 enzymes, but not E1s and E2s (21, 22). The fraction was then resolved by means of gel-filtration chromatography and the activity of the various E3 enzymes was monitored in all the fractions as described (22). To monitor the activity of E6-AP expressed in Sf9 cells, S-100 was first prepared from cell extracts and was further fractionated into Fraction II and Fraction IIA. Bacterially expressed GST-E6-11, GST-E6-16, and GST-E6-AP were induced in bacteria and purified over immobilized glutathione.
Transfection of Cells.
By using DOTAP, exponentially growing NGP cells in monolayer were stably transfected with either E6-11 or E6-16. The selective medium contained 0.8 mg/ml Geneticin. Cos cells were cotransfected transiently with human c-myc and with either E6-11 or E6-16 by using the DEAE–dextran method.
Stability of Proteins in Vivo.
The half-life of the myc proteins in the different cells was monitored in pulse-chase labeling experiments followed by immunoprecipitation, or by Western blot analysis of cell extracts following the addition of cycloheximide. The proteasome inhibitors MG132 (100 μM) or lactacystin (10 μM) were added 30 min before the beginning of the chase or to the addition of cycloheximide, and were present throughout the experiment.
Demonstration of myc-Ubiquitin Conjugates in Vivo.
To demonstrate ubiquitin conjugates of myc, NGP cells were pulse-labeled and chased in the presence of lactacystin. myc was precipitated by a specific antibody. To prevent proteolysis of conjugates by isopeptidases, cells were lysed in hot buffer as described (23). To identify the ubiquitin moiety in the conjugates, cells were treated with lactacystin, lysed, and precipitated with anti-myc antibody. The immunoprecipitate was resolved by means of SDS/PAGE, transferred onto nitrocellulose paper, and the conjugates were visualized by using anti-ubiquitin antibody (24) and enhanced chemiluminescence (ECL).
Phosphorylation of myc in Vivo.
NGP cells were pulse-labeled in the presence lactacystin and N-myc was precipitated by a specific antibody. The protein A-immobilized immunoprecipitate was treated with calf intestine alkaline phosphatase and resolved by means of SDS/PAGE.
Degradation and Conjugation Assays.
Degradation of labeled proteins was monitored essentially as described (14, 17–22). When indicated, reaction mixtures contained ≈20,000 cpm of the labeled substrate or GST-h-N-myc 1–232 (1.5 μg) and GST-E6 proteins (≈0.15 μg). Conjugates were generated in an assay similar to that used for monitoring degradation, but with the addition of ubiquitin aldehyde (0.5 μg), a specific inhibitor of certain isopeptidases (25). Reconstituted assays from purified enzymes contained 2 μg of E1, 0.3 μg of either E2-14 kDa or E2-F1, and 2.5 μg of E3α or GST-E6-AP or 25 μg of E6-AP expressing Sf9 cell extract.
N-myc⋅E6⋅E6-AP Binding Assays.
l-[35S]methionine-labeled in vitro translated (in either wheat germ or reticulocyte lysate) E6 proteins were incubated in the presence of GST-h-N-myc 1–232 immobilized to glutathione agarose. Sf9 S-100 supernatants derived from E6-AP expressing cells or cells infected with a control virus were added when indicated. The beads were washed, boiled in a sample buffer, and the proteins were resolved by means of SDS/PAGE.
RESULTS
c-myc and N-myc Are Degraded in Vivo in an ATP-Dependent Manner by Means of the Ubiquitin–Proteasome Pathway.
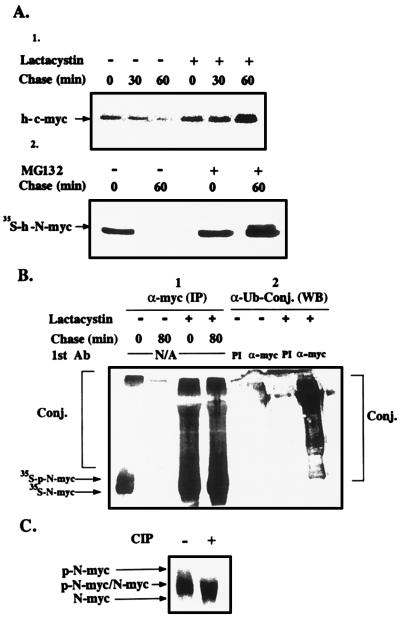
To investigate the possible role of the ubiquitin system in the degradation of c-myc and N-myc in vivo, we first examined the requirement of the proteolytic process for ATP, which is required for both conjugation and the activity of the 26S proteasome complex (10–13). In contrast, calpain-mediated degradation is energy independent. Depletion of ATP from NGP cells inhibited completely degradation of N-myc. Removal of the inhibitors and incubation of the cells in complete medium reconstituted degradation (not shown). Because lysosomal degradation also requires metabolic energy (to maintain the pH gradient across the lysosomal membrane), we examined directly the possible role of lysosomes in the proteolytic process. Addition of either chloroquine or ammonium chloride had no effect on the stability of N-myc in NGP cells (data not shown). Next, we investigated the possible involvement of the 20S proteasome in myc proteolysis. We used two inhibitors of the enzyme, the tri-peptide aldehyde derivative MG132 and the streptomyces metabolite lactacystin. As can be seen in Fig. 1A, lactacystin completely inhibits the degradation of h-c-myc. MG132 has a similar effect on the degradation of h-N-myc. MG132 and lactacystin have similar inhibitory effects on the degradation of c-myc and N-myc, respectively (not shown). Addition of the inhibitors leads to accumulation of high molecular mass derivatives (Fig. 1B-1). Western blot analysis with anti-ubiquitin serum showed that these compounds are adducts of myc with ubiquitin (Fig. 1B-2). The involvement of the ubiquitin system in the degradation of N-myc in vivo has recently been reported by Bonvini et al. (26). We noted that the inhibitors lead to accumulation of slightly slower migrating forms of the myc proteins, which are the phosphorylated forms of the proteins. Treatment of the immunoprecipitate with calf intestine alkaline phosphatase leads to “collapse” of the two phosphorylated forms of N-myc (that originate from the two initiation sites) to the respective nonphosphorylated forms (Fig. 1C; see Discussion).
Figure 1.
Effects of proteasome inhibitors on the degradation of h-c-myc and h-N-myc, on the formation of myc-ubiquitin conjugates, and on the stabilization of phosphorylated myc in vivo. (A) Proteasome inhibitors stabilize c-myc and N-myc in vivo. [Upper (1)] Cos cells that were transiently transfected with c-myc were treated with cycloheximide and harvested at the indicated times. Dimethyl sulfoxide or lactacystin were added 30 min before the addition of cycloheximide. c-myc was detected by means of Western blot analysis and ECL. [Lower (2)] A similar experiment was carried out in the N-myc expressing NGP cells, except that MG132 was used as a proteasome inhibitor and the fate of the protein was monitored in a pulse-chase experiment and immunoprecipitation following metabolic labeling. (B) Proteasome inhibitors lead to accumulation of ubiquitin-myc conjugates in vivo. [Left (1)] NGP cells were pulse-labeled and chased in the presence of lactacystin or dimethyl sulfoxide. Labeled myc and myc conjugates were detected by means of phosphorimaging following immunoprecipitation with anti-myc antibody. [Right (2)] NGP cells were treated with lactacystin or dimethyl sulfoxide for 60 min, lysed, and the lysates were then treated with anti-myc or preimmune (PI) antibody. Immunoprecipitates were resolved by means of SDS/PAGE, and following transfer of the proteins to nitrocellulose paper conjugates were detected by anti-ubiquitin antibody and by ECL. (C) myc is phosphorylated in vivo. NGP cells were pulse-labeled in the presence of lactacystin. The immunoprecipitated protein, still bound to immobilized protein A, was treated, when indicated, with calf intestine alkaline phosphatase and resolved by means of SDS/PAGE. N-myc was visualized following exposure to a PhosphorImager screen. p-N-myc denotes phosphorylated N-myc.
Cell-Free System for Conjugation and Degradation of myc Proteins: Involvement of the High-Risk HPV E6-16 in the Process.
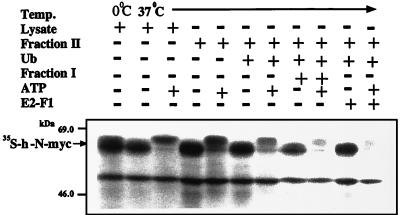
To identify and characterize the conjugating enzymes and ancillary proteins involved in ubiquitin-mediated proteolysis of myc, we reconstituted the system in vitro. As can be seen in Fig. 2, incubation of N-myc in the presence of complete reticulocyte lysate and ATP results in significant degradation of the protein. ATP-supplemented Fraction II does not promote degradation, but the addition of ubiquitin stimulates degradation significantly. Notably, the addition of Fraction I stimulates degradation even further. Fraction I contains several E2 enzymes, among them E2-F1 (21). Indeed, addition of purified E2-F1 faithfully reproduced the stimulatory effect of Fraction I. As can be seen clearly, N-myc is phosphorylated during incubation in the presence of Fraction II and ATP (see above and in Discussion).
Figure 2.
Degradation of h-N-myc in a cell-free reconstituted system requires ubiquitin and ATP, and is stimulated by Fraction I and E2-F1. Degradation of h-N-myc was monitored in a cell-free reconstituted system in the presence of reticulocyte lysate or Fraction II supplemented with ubiquitin and Fraction I or E2-F1 as indicated. ATP and the ATP-regenerating system or a system that depletes ATP were added as indicated.
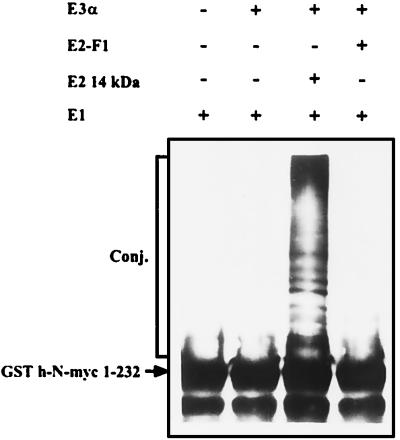
The finding that both Fraction II, which contains several species of E2s, and E2-F1, which is contained in Fraction I, can stimulate degradation of N-myc, raised the assumption that the proteolytic process is mediated by at least two E2 enzymes, and possibly two E3s. Fraction II contains all known E3 enzymes, in addition to several E2 enzymes, including E2-14 kDa, which along with E3α is involved in targeting substrates by means of the “N-end rule” pathway (13). As can be seen in Fig. 3, E2-14 kDa and E3α can efficiently conjugate h-N-myc, suggesting that E2-14 kDa/E3α can act also in the cell as one route for myc degradation. As expected, E3α does not work in concert with E2-F1.
Figure 3.
E2-14 kDa- and E3α-dependent conjugation of h-N-myc. Conjugation of h-N-myc was monitored in the presence of the indicated enzymes. Proteins were detected by means of Western blot analysis by using anti-N-myc antibody.
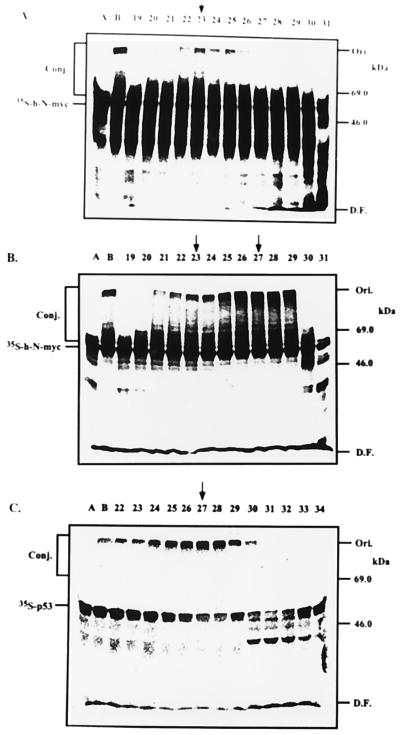
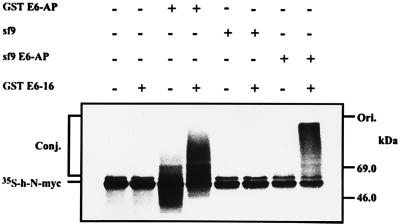
Because the addition of E2-F1 stimulated the degradation of N-myc beyond the level observed in the presence of Fraction II alone (Fig. 2), and because E2-F1 does not catalyze E3α-mediated conjugation (Fig. 3), it was clear that this enzyme must act in concert with another species E3. To identify this enzyme, we monitored E3 activity in gel filtration-resolved Fraction IIA that contains several known species of E3s (21, 22). As can be seen in Fig. 4A, a peak of E3 activity is observed in Fraction 23, corresponding to molecular mass of ≈280 kDa. This enzyme was characterized further by additional chromatography steps and it appears to be similar or identical to the c-Fos conjugating E3 (22). Because E2-F1 catalyzes conjugation of p53 in a reaction that involves the high-risk HPV oncoprotein E6-16 and the E3 enzyme E6-AP (15), we decided to examine the possible role of E6 in the proteolysis of myc proteins. As can be seen in Fig. 4B, addition of E6-16 to the reaction mixtures containing the different fractions of the gel filtration-resolved Fraction IIA stimulated conjugation severalfold. This E6-dependent activity is mediated however by a different E3 enzyme, as it peaks in Fraction 27, in a molecular mass region of ≈200 kDa. To identify the E6-dependent E3 enzyme, we monitored conjugation of p53 in the same fractions in the presence of E6. As can be seen in Fig. 4C, the p53-conjugating activity peaks also in Fraction 27, strongly suggesting that the E3 that conjugates N-myc in the presence of E6 is also E6-AP. To demonstrate directly the involvement of E6-AP N-myc conjugation, we reconstituted a cell-free system from purified enzymes. As can be seen in Fig. 5, either bacterially or baculovirus expressed E6-AP can promote conjugation of h-N-myc in the presence, but not in the absence, of E6. At this point, it was important to show that E6 can stimulate not only the conjugation but also the degradation of the myc proteins. As can be seen in Fig. 6 A and B, addition of E6-16 to a crude extract stimulates conjugation of the myc proteins severalfold and generates high molecular mass conjugates. Concomitantly, degradation is also accelerated (Fig. 6 C and D). In contrast, addition of the low-risk oncoprotein E6-11 did not affect conjugation (not shown) and degradation (Fig. 6D) of the myc proteins. The conjugation and degradation observed in the absence of E6 probably reflects the basal activity of the two pairs of the conjugating enzymes, E2-14 kDa/E3α and E2-F1/E3-c-Fos.
Figure 4.
E6-independent and -dependent ubiquitin ligases are involved in h-N-myc conjugation. Fraction IIA was resolved over a Superdex 200 gel filtration column (connected to a fast protein liquid chromatography apparatus; Pharmacia), and the different fractions were assayed for E3 activity in the presence of purified E1 and E2-F1. Lanes A, reactions carried out in the presence of E1 and E2-F1. Lanes B, reactions carried out in the presence of E1, E2-F1, and crude Fraction IIA (10 μg). Numbers denote reactions that were carried out in the presence of E1, E2-F1, and an aliquot (2 μl) from the appropriate column fractions. Ubiquitin conjugates, the labeled substrates, and molecular mass markers are indicated. Arrow pointing to fraction 23 indicates peak activity of E6-independent ligase, whereas arrows pointing to fraction 27 indicate peak activity of E6-dependent ligase. (A) Conjugation of h-N-myc in the absence of E6. (B) Conjugation of h-N-myc of the presence of E6-16. (C) Conjugation of p53 in the presence of E6-16.
Figure 5.
Reconstitution of conjugation of h-N-myc from purified conjugating enzymes. h-N-myc translated in vitro in wheat germ extract that lacks E6-AP was incubated for 10 min at room temperature in the presence of 10 mM N-ethylmaleimide to inactivate the endogenous E1 and E2 enzymes. DTT was added to a final concentration of 6 mM to neutralize the N-ethylmaleimide. Conjugation reactions were carried out in the presence of purified E1, E2-F1, and either GST-E6-AP or baculovirus-expressed E6-AP and GST-E6-16 as indicated.
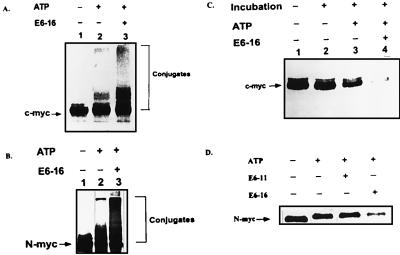
Figure 6.
E6-16-dependent conjugation and degradation of h-c-myc and h-N-myc. Conjugation (A and B) and degradation (C and D) of l-[35S]methionine-labeled h-c-myc (A and C) and h-N-myc (B and D) were monitored in crude reticulocyte lysate in the presence or absence of E6 proteins as indicated.
It should be noted that the addition of Max does not have any effect on the conjugation and degradation of Myc either in the absence or the presence of E6 (not shown). Max itself is stable.
E6-16 Accelerates Degradation of myc Proteins in Vivo.
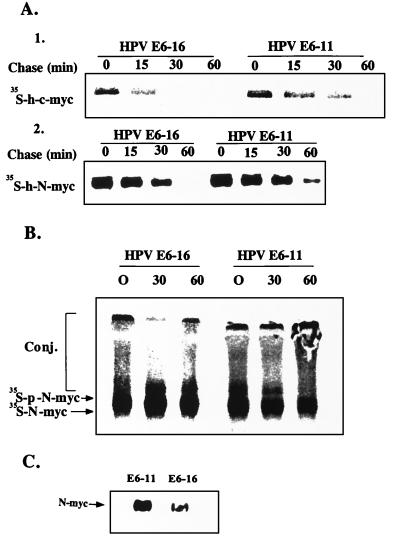
To investigate the role of E6-16 in the proteolytic process in vivo, we expressed transiently in Cos cells c-myc with either the high-risk E6-16 protein or with its low-risk counterpart, E6-11. In parallel, we tested the effect of the two proteins following their stable transfection into the neuroblastoma cell line NGP that express constitutively N-myc. As can be seen in Fig. 7A, expression of E6-16 with either c-myc or N-myc results in a significant shortening of the half-life of the proteins. Quantitation of data from six different experiments revealed that the average shortening of the t1/2 was 2.7-fold (1.9- to 4.0-fold in the different experiments). In contrast, expression of E6-11 does not affect the stability of the myc proteins. Interestingly, lactacystin inhibits both the basal (in the presence of E6-11) and the accelerated (in the presence of E6-16) degradation of N-myc (Fig. 7B), strongly suggesting that the two processes are mediated by the ubiquitin system. In agreement with these findings, the steady-state level of N-myc in the E6-16-expressing NGP cells is ≈3-fold lower compared with cells expressing E6-11 (Fig. 7C).
Figure 7.
Effect of E6 proteins and lactacystin on the degradation and steady-state level of h-N-myc and h-c-myc in vivo. (A) Effect of E6-16 and E6-11 on the degradation of myc proteins in vivo. Cos cells [Upper (1)] were cotransfected transiently with h-c-myc and either E6-11 or E6-16, and NGP cells [Lower (2)] were stably transfected with either E6-11 or E6-16. Stability of the myc proteins was determined in the cells in a pulse-chase labeling experiment followed by immunoprecipitation and detection of the labeled myc proteins by means of phosphorimaging. (B) Effect of lactacystin on basal and E6-16-induced degradation of N-myc. NGP cells were pulse-labeled and chased for the indicated times in the presence of lactacystin. N-myc was immunoprecipitated, resolved by means of SDS/PAGE, and visualized by PhosphorImager. (C) Steady-state level of N-myc in NGP cells in the presence of E6-11 and E6-16. The level of N-myc was monitored in NGP cells stably transfected with either E6-11 or E6-16 by using Western blot analysis and ECL. p-N-myc denotes phosphorylated N-myc.
E6 Binds to N-myc in an E6-AP-Dependent Manner.
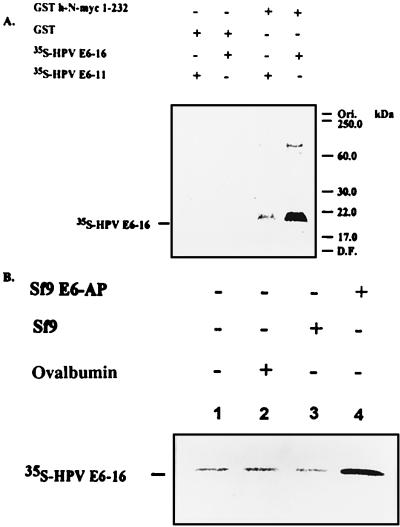
Degradation of p53 involves formation of a p53⋅E6⋅E6-AP targeting complex that enables, most probably, efficient ubiquitination of p53. We tested whether E6 promotes the generation of a similar complex with myc as well. As can be seen in Fig. 8A, immobilized N-myc can bind E6-16, but not E6-11. The reaction requires a component that is contained in reticulocyte Fraction II that was added along with the labeled E6 proteins (that were translated in reticulocyte lysate). To identify the component in Fraction II involved in the formation of the complex, E6-16 was translated in wheat germ extract that does not contain E6-AP. As can be seen in Fig. 8B, addition of recombinant E6-AP expressed in Sf9 cells was necessary to bind the labeled E6-16. Cytosol derived from cells that do not express E6-AP was inactive.
Figure 8.
E6-AP- and E6-16-dependent formation of a ternary targeting complex with h-N-myc. (A) Immobilized GST-h-N-myc 1–232 was incubated in the presence of E6-11 and E6-16 translated in vitro in reticulocyte extract in the presence of l-[35S]methionine. The beads were washed and the labeled proteins were detected following SDS/PAGE by using fluorography. (B) E6-16 protein was translated in wheat germ that lacks E6-AP and was incubated in the presence of Sf9 cell extract that express E6-AP or an extract from untransfected cells. Immobilized GST h-N-myc 1-232 was present in all essays.
DISCUSSION
We have shown that c-myc and N-myc are targeted for degradation by the ubiquitin system. We noted that the proteasome inhibitors lead to accumulation of a phosphorylated form of the myc proteins that is also formed during incubation of myc in the presence of ATP and Fraction II (Figs. 1, 2, and 6). Initial kinetic analysis of degradation data reveals that the nonphosphorylated form is converted first to the modified protein, which is subsequently degraded (not shown). It is possible that a specific phosphorylation signals these proteins for degradation, as is the case with other substrates of the ubiquitin system, IκBα for example.
In our attempts to identify the targeting enzymes involved, we have shown that in a reconstituted cell-free system, N-myc can be conjugated by at least two pairs of E2/E3 enzymes, E2-14 kDa/E3α and E2-F1/E3-c-Fos (Figs. 2–4 and data not shown; because E3-Fos was not purified to homogeneity, it is not clear whether the myc-E3 is identical or only similar to the c-Fos E3). The finding that E2-14 kDa and E3α conjugate N-myc prompted us to study the possibility that the protein is targeted by means of the N-end rule pathway. Specific dipeptides that inhibit the pathway (Arg–Ala and Phe–Ala; ref. 20) failed to inhibit degradation of the protein in vitro. Also, theoretical considerations concerning the identity of the N-terminal residue rule out the possibility that it is a “destabilizing” residue (27). Thus, E2-14 kDa/E3α must target N-myc by means of a signal that is distinct from its N-terminal residue. The fact that E2-F1 stimulates degradation beyond the level observed with Fraction II alone suggests that the two E3 enzymes, E3α and E3-Fos, target myc by means of two distinct signals. It has been shown previously that certain substrates of the ubiquitin system can be targeted by distinct pairs of conjugating enzymes recognizing different targeting signals (16, 17). It is possible that the apparently functionally redundant mechanisms were evolved evolutionarily as a safeguard against inactivating mutations in key metabolic pathways.
The finding that E6-16 stimulates the degradation of the myc proteins is rather surprising. Whereas the rationale behind the removal of p53 by the viral oncoprotein is readily understood, it is more difficult to envision the evolutionary mechanisms that underlie the removal of myc by E6. It is possible that the virus developed a defense mechanism against myc-induced apoptosis. Under certain conditions, when cells cannot proliferate, forced expression of myc leads to reentry into the cell cycle and to apoptosis (5, 6). It is possible that E6, by targeting myc for degradation, prevents the apoptotic process and ascertains continuous propagation of the viral infection.
Of note is that Max, which heterodimerizes with myc in the cell, does not affect the E6-induced degradation of myc. It is not clear however whether in the cell myc is degraded as an integral part of the complex or whether it has to dissociate first to be recognized by the ubiquitin conjugating machinery.
An interesting finding is that in invasive genital cancers, the E6-16 and E6-18 genes have been found to be integrated in chromosome band 8q24.1 to which the c-myc has been mapped. In three of four reported cases, the protooncogene was found to be structurally altered and/or overexpressed (28). Overexpression of c-myc was found in certain cases of uterine cervical carcinoma and was associated with greater incidence of early relapse and invasiveness (29). This expression of myc was distributed equally among HPV-positive and -negative patients. These findings support the hypothesis that the integration plays a role in tumor progression by means of activation of the cellular oncogene. The apparently paradoxical findings of HPV-dependent activation of c-myc and E6-dependent targeting of myc for degradation may reflect the complex roles of the myc proteins in determining the various fates of the cell under different pathophysiological conditions: proliferation, malignant transformation, cell cycle arrest, differentiation, and apoptosis. Activation or targeting for degradation may be cell-specific phenomena that occur under distinct environmental conditions.
Acknowledgments
We thank Dr. Martin Scheffner from the DKFZ (Germany) for the E6 plasmids; Drs. Jon Huibregtse (Rutgers University), Martin Scheffner, and Peter Howley (Harvard Medical School) for the E6-AP plasmids; and Dr. Garrett Brodeur (Children’s Hospital, Philadelphia) for the NGP cell line. This research was supported by grants from the German–Israeli Foundation for Scientific Research and Development; Israel Science Foundation, founded by the Israeli Academy of Sciences and Humanities—Centers of Excellence Program; U.S.–Israel Binational Science Foundation; Israeli Ministry of Science; U.K.–Israel Science and Technology Research Fund; Foundation for Promotion of Research at the Technion; and a research grant administered by the vice president of the Technion for research (to A.C.). C.K. holds the Jules J. Mallon Chair of Biochemistry and was supported by grants from the Israel Cancer Society and La Fondation Raphael et Regina Levy. A.L.S. was supported by a grant from the National Institutes of Health.
ABBREVIATIONS
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-carrier protein or ubiquitin-conjugating enzyme (Ubc)
- E3
ubiquitin-protein ligase
- ECL
enhanced chemiluminescence
- E6-AP
E6-associated protein
- HPV
human papillomavirus
- GST
glutathione S-transferase
References
- 1.Luscher B, Eisenman R N. Genes Dev. 1990;4:2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- 2.Landschulz W H, Johnson P F, McKnight S L. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 3.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 4.Spencer C A, Groudine M. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 5.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 6.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 7.Marcu K B, Bossone S A, Patel A J. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 8.Dani C, Blanchard J-M, Piechaczyk M, El-Sabouty S, Marty L, Jeanteur P. Proc Natl Acad Sci USA. 1984;81:7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsay G, Stanton L, Schwab M, Bishop M. Mol Cell Biol. 1986;6:4450–4457. doi: 10.1128/mcb.6.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershko A. Trends Biochem Sci. 1996;21:445–449. doi: 10.1016/s0968-0004(96)10054-2. [DOI] [PubMed] [Google Scholar]
- 11.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 12.Ciechanover A, Schwartz A L. Proc Natl Acad Sci USA. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varshavsky A. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 14.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz A L, Ciechanover A. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 15.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 17.Gonen H, Stancovski I, Shkedy D, Hadari T, Bercovich B, Bengal E, Mesilaty S, Abu-Chatoum O, Schwartz A L, Ciechanover A. J Biol Chem. 1996;271:302–310. doi: 10.1074/jbc.271.1.302. [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A, DiGiuseppe J A, Bercovich B, Orian A, Richter J D, Schwartz A L, Brodeur G M. Proc Natl Acad Sci USA. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershko A, Heller H, Elias S, Ciechanover A. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 20.Reiss Y, Hershko A. J Biol Chem. 1990;265:3685–3690. [PubMed] [Google Scholar]
- 21.Blumenfeld N, Gonen H, Mayer A, Smith C E, Siegel N R, Schwartz A L, Ciechanover A. J Biol Chem. 1994;269:9574–9581. [PubMed] [Google Scholar]
- 22.Stancovski I, Gonen H, Orian A, Schwartz A L, Ciechanover A. Mol Cell Biol. 1995;15:7106–7116. doi: 10.1128/mcb.15.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strous G J, van Kerkhof P, Govers R, Ciechanover A, Schwartz A L. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- 24.Hershko A, Eytan E, Ciechanover A, Haas A L. J Biol Chem. 1982;257:13964–13970. [PubMed] [Google Scholar]
- 25.Hershko A, Rose I A. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonvini P, Nguyen P, Trepel J, Neckers L M. Oncogene. 1998;16:131–139. doi: 10.1038/sj.onc.1201625. [DOI] [PubMed] [Google Scholar]
- 27.Boissel J P, Casper T J, Bunn H F. J Biol Chem. 1988;263:8443–8449. [PubMed] [Google Scholar]
- 28.Couturier J, Sastre-Garau X, Schneider-Maunoury S, Labib A, Orth G. Virology. 1991;65:4534–4538. doi: 10.1128/jvi.65.8.4534-4538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riou G, L, Favre M, Jeannel D, Bourhis J, Orth G. J Natl Cancer Inst. 1992;84:1525–1526. doi: 10.1093/jnci/84.19.1525. [DOI] [PubMed] [Google Scholar]