Abstract
IL-4, an anti-inflammatory cytokine, inhibits osteoclast differentiation, but the basis of this effect has been unclear. Osteoclastogenesis requires activation of RANK, which exerts its biologic effect via activation of NF-κB. NF-κB activation is manifested by nuclear translocation and binding to DNA, events secondary to phosphorylation and dissociation of IκBα. It is shown here that IL-4 reduces NF-κB nuclear translocation by inhibiting IκB phosphorylation, thus markedly inhibiting NF-κB DNA binding activity and blocking osteoclastogenesis entirely. Residual translocation of NF-κB in the presence of IL-4, however, suggests that nuclear mechanisms must primarily account for inhibition of NF-κB DNA binding and blockade of osteoclastogenesis. To address this issue, this study examined whether IL-4–induced STAT6 transcription factor blocks NF-κB transactivation. The results show that excess unlabeled consensus sequence STAT6, but not its mutated form, inhibits NF-κB binding. Furthermore, exogenously added STAT6 protein inhibits NF-κB/DNA interaction. Further supporting a role for STAT6 in this process are the findings that IL-4 fails to block osteoclastogenesis in STAT6–/– mice but that this blockade can be restored with addition of exogenous STAT6. Thus, IL-4 obliterates osteoclast differentiation by antagonizing NF-κB activation in a STAT6-dependent manner.
Introduction
Osteoclast differentiation and function are required for normal bone resorption and remodeling (1, 2). The mature osteoclast polykaryon arises from marrow-residing precursor cells of hematopoietic origin, believed to be monocytes/macrophages (1–3). Early studies have shown that cocultures of these precursor cells with stromal/osteoblastic cells in the presence of hormonal steroids led to differentiation of mature osteoclast-like cells capable of resorbing bone (4–6). Recent advances revealed that factors produced by osteoblastic cells, such as M-CSF and receptor activator of NF-κB ligand (RANKL), are the factors required for macrophage differentiation into mature bone-resorbing osteoclasts (7–10). Although M-CSF is a prerequisite for macrophage and osteoclast survival, RANKL appears to be the sole factor essential for acquiring the osteoclastic phenotype. RANKL, a transmembrane protein, and its cleaved soluble form have been shown to be effective in inducing osteoclast differentiation through binding to its receptor RANK in osteoclast progenitors (11, 12). Although details of the intracellular signal transduction pathway after RANK/RANKL interaction are not entirely in hand, activation of the transcription factors NF-κB and AP-1 has been proposed (7, 12–15). NF-κB is essential for osteoclastogenesis, and inactivation of components of this transcription factor led to osteopetrosis due to impaired osteoclast formation (16, 17).
Inactive NF-κB is normally present in the cytoplasm complexed with its inhibitory protein IκB (18–20). A variety of extracellular stimuli activate a cascade of kinases including IκB kinases (IKKs), the MAP kinase MEKK-1, and c-src, leading to phosphorylation of IκB and to its dissociation from NF-κB (21–26). Liberated NF-κB translocates in a dimeric form to the nucleus, binds to DNA response elements, and activates basal transcription of target genes (18, 19).
The osteoclastogenic process is regulated by immune cell–derived cytokines. The role of lymphocyte-derived cytokines that impact bone metabolism under physiological and pathological conditions has been steadily investigated in the past few years (27–29). Although cytokines secreted by Th1-type lymphocytes, such as RANKL and TNF, induce osteoclastogenesis, those produced by Th2 cells, such as IL-4, IL-10, and IL-13, have been shown as potent inhibitors of the osteoclastogenic process (6, 27, 28, 30–32). However, the molecular mechanisms by which these anti-inflammatory cytokines curtail osteoclast differentiation remain obscure. IL-4 is a pleiotropic immunomodulatory cytokine produced by Th2 lymphocytes, mast cells, and eosinophils (33, 34). The cytokine promotes immunological responses, and its levels are elevated in tissues succumb to chronic inflammatory diseases (35–37). IL-4 ligation to its receptor activates protein tyrosine kinases of the Janus kinase (JAK) family, which in turn leads to phosphorylation, dimerization, and nuclear translocation of the transcription factor STAT6, a member of the family of signal transducers and activators of transcription (38–41). In the nucleus, STAT6 binds to specific regions of promoters of target genes. Several reports described STAT6 as an essential component of Th2 lymphocyte responses and critical for many IL-4 functions (42).
Previous reports have described the osteoclast inhibitory role of IL-4 (29, 31, 43–45). Just how this cytokine abrogates osteoclastogenesis remains unclear. This report explores the molecular steps of IL-4 inhibition of RANKL-mediated events throughout osteoclastogenesis. The results indicate that IL-4, via targeting osteoclast precursors, exerts its inhibitory effect by targeting NF-κB activation. IL-4 activates STAT6, which antagonizes NF-κB DNA-binding activity. Therefore, IL-4–activated STAT6 may be acting as a transcriptional repressor of NF-κB–activated genes.
Methods
Reagents.
Polyclonal anti-IκBα and anti–NF-κB antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, California, USA). Recombinant murine IL-4 and M-CSF were purchased from R&D Systems Inc. (Minneapolis, Minnesota, USA). M. Tondravi (Red Cross, Rockville, Maryland, USA) generously provided RANKL. The ECL kit was obtained from Pierce Chemical Co. (Rockford, Illinois, USA). All other chemicals were obtained from Sigma Chemical Co. (St. Louis, Missouri, USA). The primers used were: OPG forward primer, 5′-ACC ACT ACT ACA GAC AGC-3′; and OPG reverse primer, 5′-AGG AGA CCA AAG ACA CTG CA-3′.
Animals.
C3H/HeN males were purchased from Harlan Industries (Indianapolis, Indiana, USA). STAT6 knockout and BALB/c control mice were purchased from The Jackson Laboratory (Bar Harbor, Maine, USA).
Cell culture.
Bone marrow macrophages (BMMs) were isolated from whole bone marrow of 4- to 6-week-old mice and incubated in tissue culture plates, at 37°C in 5% CO2, in the presence of 10 ng/ml M-CSF (46, 47). After 24 hours in culture, the nonadherent cells were collected and layered on a Ficoll-Hypaque gradient. Cells at the gradient interface were collected and plated in α-MEM, supplemented with 10% heat-inactivated FBS at 37°C in 5% CO2 in the presence of 10 ng/ml M-CSF, and plated according to each experimental conditions.
Immunopurification of mouse monocytes/macrophages.
Ficoll-Hypaque gradient-purified macrophages were further purified with CD11b (PharMingen, San Diego, California, USA) antibody and Dynabeads (Dynal Inc., Lake Success, New York, USA) following the manufacturer’s instructions.
Osteoclast generation.
Purified marrow macrophages were cultured at 1 × 106 cells/ml in the presence of 10 ng/ml M-CSF and 20 ng/ml RANKL for 4 days. Cultures were supplemented with M-CSF and RANKL on day 2 of culture.
Immunoblotting.
Total cell lysates were boiled in the presence of 2xSDS-sample buffer (0.5 M Tris-HCl [pH 6.8], 10% [wt/vol] SDS, 10% glycerol, 0.05% [wt/vol] bromophenol blue, distilled water) for 5 minutes and subjected to electrophoresis on 8–12% SDS-PAGE (48). Proteins were transferred to nitrocellulose membranes using a semidry blotter (Bio-Rad Laboratories Inc., Richmond, California, USA) and incubated in blocking solution (10% skim milk prepared in PBS containing 0.05% Tween-20), to reduce nonspecific binding. Membranes were washed with PBS/Tween buffer and exposed to primary antibodies (1 hour at room temperature for as long as overnight at 4°C), washed again four times,and incubated with the respective secondary HRP-conjugated antibodies (1 hour at room temperature). Membranes were washed extensively (5 × 15 min), and an ECL detection assay was performed following the manufacturer’s directions.
Electrophoretic mobility shift assay.
Nuclear fractions were prepared as described previously (26, 49). In brief, monolayers of BMMs grown in a 100-mm2 tissue culture dish were washed twice with ice-cold PBS. Cells were lifted from the dish by treating with 5 mM EDTA and 5 mM EGTA in PBS. Cells were then resuspended in hypotonic lysis buffer A (10 mM HEPES [pH 7.8], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol 0.5 mM AEBSF, and 5 μg/ml Leupeptin) and incubated on ice for 15 minutes, and NP-40 was added to a final concentration of 0.64%. Nuclei were pelleted, and the cytosolic fraction was carefully removed. The nuclei were then resuspended in nuclear extraction buffer B (20 mM HEPES[pH 7.8], 420 mM NaCl, 1.2 mM MgCl2, 0.2 mM EDTA 25% glycerol, 0.5 mM dithiothreitol, 0.5 mM AEBSF, 5 μg/ml Pepstatin A, and 5 μg/ml Leupeptin), vortexed for 30 seconds, and rotated for 30 minutes in 4°C. The samples were then centrifuged, and the nuclear proteins in the supernatant were transferred to fresh tubes and protein content was measured using a standard BCA kit (Pierce Chemical Co.). Nuclear extracts (10 μg) were incubated with an end-labeled double-stranded oligonucleotide probe containing the sequence 5′- AAA CAG GGG GCT TTC CCT CCT C -3′ (50) derived from the κB3 site of the TNF promoter. Commercial STAT6 consensus oligo 5′- GTA TTT CCC AGA AAA GGA AC –3’ and mutant oligo 5′- GTA TTT Cgg ttA AAA GGA AC –3’ (Santa Cruz) were also used. The reaction was performed in a total of 20 μl of binding buffer (20 mM HEPES [pH 7.8], 100 mM NaCl, 0.5 mM dithiothreitol, 1 μg poly dI-dC, and 10% glycerol) for 30 minutes at room temperature. Samples were then fractionated on a 4% polyacrylamide gel and visualized by exposing dried gel to film.
pTAT construct and protein coupling.
STAT6 construct was cloned into the pTAT-HA bacterial expression vector described by Nagahara, Dowdy, and colleagues previously (51–53) that contains a six-histidine tag, for easy purification, HA tag for detection followed by the TAT transduction domain, and finally, the STAT6 sequence. The resultant plasmid, pTAT-STAT6, was transformed into the DH5α strain of Escherichia coli. The transformants were screened initially by restriction enzyme mapping. A recombinant containing the correct restriction fragments was then sequenced on both strands. This plasmid was then used to express the TAT-coupled STAT6 in the BL21 (DE3) strain of E. coli. After a 4- to 6-hour induction, the cells were sonicated in 8 M urea and the TAT-coupled STAT6 was purified on a nickel-Sepharose column (QIAGEN Inc., Valencia, California, USA), and then applied to an ionic exchange column (Mono Q) in 4 M urea. To shock misfold the protein, the ionic exchange column was switched in one step from 4 M urea to aqueous buffer (20 mM HEPES). STAT6 was eluted by stepping from 50 mM to 1 M NaCl followed by desalting on a PD-10 column (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) into PBS and frozen in 10% glycerol at –80°C. pTAT-coupled and misfolded proteins remain highly concentrated and resistant to freeze-thaw denaturation and readily enter the cells upon incubation. Once in hand, the coupled TAT-STAT6 was added to cultured cells without the aid of transfection agents.
Results
IL-4 blocks basal RANKL-mediated osteoclastogenesis by BMMs.
To identify whether IL-4 abrogates osteoclastogenesis by targeting osteoclast precursor cells directly, pure populations of marrow macrophages were incubated with RANKL and M-CSF in the absence or presence of 10 ng/ml murine IL-4. Osteoclast differentiation was assessed using tartrate-resistant acid phosphatase (TRAP) assay. The results indicate that IL-4 blocks RANKL-mediated osteoclastogenesis (Figure 1), evident by complete lack of multi- and mononucleated TRAP-expressing cells in a pattern similar to controls. The cytokine totally inhibits osteoclast formation at concentrations as low as 0.1 ng/ml (data not shown). The apparent increase in non-TRAP cells in the IL-4–treated conditions represents an increase in cell adherence and spreading, as IL-4 upregulates the macrophages β3 integrin (54). Because the cell population used includes 5–10% contaminating cells, primarily stromal cells, the IL-4 inhibitory effect on immunopurified monocytes/macrophages was tested. In this exercise, cells were purified using magnetic bead-conjugated CD11b (MAC-1) antibody, resulting in an approximately 98% pure population of monocytes/macrophages. IL-4 inhibition of osteoclast formation by these cells (data not shown) was identical to the results shown in Figure 1.
Figure 1.
IL-4 blocks RANKL-mediated osteoclastogenesis by bone marrow macrophages. Osteoclast precursor cells were isolated from the bone marrow of 4- to 6-week-old mice as described in Methods. Pure (∼90%) marrow macrophages were plated in 48-well plates at 1 × 106 cells/ml using α-MEM supplemented with 10% heat-inactivated (HI) FCS and 10 ng/ml M-CSF. Cells were treated with PBS, 10 ng/ml mIL-4 for 30–60 minutes, 20 ng/ml soluble RANKL for 4 days, or a combination of IL-4 and RANKL (as shown). Cultures were placed at 37°C in a 5% CO2 incubator and supplemented with an additional dose of M-CSF and RANKL on the third day of culture. Osteoclasts developed on days 4–5, after which they were washed, fixed, and stained for TRAP activity following manufacturers’ directions. TRAP-positive (purple) mono- and multinucleated large cells are osteoclasts and their committed precursors. The average number of osteoclasts in RANKL-treated cultures was 182 ± 22/cm2 compared with no osteoclasts in all other conditions. Results represent average number of quadruplicate wells from three independent experiments. ×20 taken by light microscope.
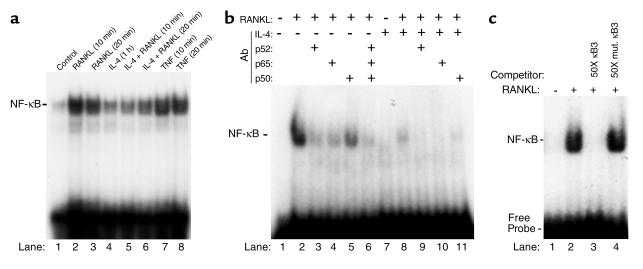
IL-4 inhibits NF-κB DNA binding activity.
The previous experiment has established that IL-4 blocks osteoclastogenesis by targeting the macrophage precursor cells. Thus, the next set of experiments was designed to delineate the molecular mechanism(s) by which IL-4 exerts its inhibitory effect. The osteoclastogenic factor RANKL transmits its signals via binding to its transmembranal receptor RANK. This interaction leads to activation of the transcription factor NF-κB, deletion of which led to osteoclast depletion and osteopetrosis. Thus, the question of whether IL-4 impacts RANKL activation of NF-κB was explored. To this end, marrow macrophages were cultured to confluence, incubated with IL-4 for 1 hour followed by addition of 20 ng/ml RANKL. TNF-treated cell extracts were included as positive controls. Electrophoretic mobility shift assay (EMSA) of nuclear extracts indicates that IL-4 pretreatment significantly inhibits RANKL-induced NF-κB activation (Figure 2a). Densitometric analysis revealed an 84% and 71% inhibition of RANKL-induced NF-κB by IL-4 at 10 and 20 minutes, respectively. To identify further the composition of the NF-κB shifted band, the EMSA reaction was performed in the presence of p50, p52, and p65 NF-κB antibodies. The data shown in Figure 2b indicate that at least all three NF-κB subunits are present in the NF-κB complex, evident by reduced NF-κB binding activity in the presence of each antibody and by depletion of the shifted band in the presence of combined addition of the NF-κB antibodies. P52 and p65 subunits, however, dominate this complex. IL-4 significantly inhibited NF-κB band shift (lane 8) and p52, p65 antibodies further abolished this negligible NF-κB binding (lanes 9 and 10). Super-shifted bands were not present because NF-κB antibodies blocked binding to DNA.
Figure 2.
IL-4 inhibits NF-κB DNA binding activity. (a) Pure population of bone marrow macrophages was grown to confluence for 4 days in the presence of M-CSF. Cells were then treated with 10 ng/ml mIL-4 for 1 hour followed by 20 ng/ml RANKL as indicated. At the end of treatment, cells were harvested, nuclear extracts were prepared, and NF-κB EMSA was performed as described in Methods. Similar results represent three independent experiments. Incomplete inhibition of RANKL-induced NF-κB by IL-4 in lanes 5 and 6 correlates with basal expression of NF-κB under IL-4–treated conditions (lane 4), which does not support osteoclastogenesis. Extracts from 10- and 20-minute TNF-treated cells (10 ng/ml) were included as a positive control. (b) Identity of the NF-κB band was confirmed by binding with various NF-κB subunit antibodies (1 μl/reaction) as indicated or by a representative nonimmune IgG (lane 2). (c) Specificity of NF-κB binding was assessed by the addition of 50-fold (50×) unlabeled or mutated oligonucleotides, resulting in complete displacement or lack of displacement of the labeled probe, respectively.
Binding specificity was further established by competing the NF-κB band with excess unlabeled oligonucleotide and failure of such competition with the mutated form of the oligonucleotide (Figure 2c).
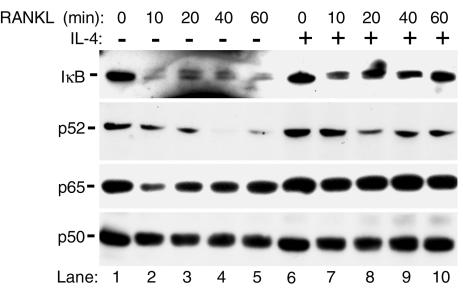
IL-4 inhibits IκB phosphorylation and NF-κB nuclear translocation.
Under naive conditions, inactive NF-κB is found in the cytosol bound to IκB. Phosphorylation and release of IκB facilitate NF-κB nuclear translocation and activation. Thus, it was reasoned that IL-4 inhibition of NF-κB is due to inhibition of IκB/NF-κB dissociation. Examination of IκB in macrophages revealed that IL-4 prevents RANKL induction of IκB phosphorylation and degradation (Figure 3). Next, cytoplasmic to nuclear translocation of NF-κB in the absence or presence of IL-4 was examined. The data shown in Figure 3 indicate that RANKL induces nuclear translocation of p52 and p65 subunits within 10 minutes after exposure, reflected by decreasing cytoplasmic levels of these two proteins with no evident change in p50 subunit level. More importantly, and consistent with IL-4-inhibition of IκB phosphorylation/degradation, IL-4 when added prior to RANKL totally prevents the decrease in cytoplasmic p65 and moderately inhibited that of p52.
Figure 3.
IL-4 inhibits IκB phosphorylation and NF-κB nuclear translocation. Cells were grown as described for Figure 2 and treated with RANKL for the amounts of time indicated in the absence or presence of pretreatment with IL-4. Cells were then lysed, and cytoplasmic and nuclear fractions were separated. Cytosols were analyzed for IκB, p50, p52, and p65 NF-κB by immunoblots. Anti-IκB antibody raised against the carboxy terminus of the protein that recognizes both native and phosphorylated IκB was used. Results are representative of two independent experiments.
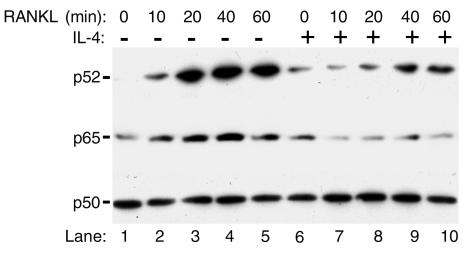
Residual levels of NF-κB proteins are present in nuclei in the presence of IL-4.
Despite significant inhibition of NF-κB nuclear translocation by IL-4, careful examination of the data in Figure 3 indicates that IL-4 did not affect p50 NF-κB and failed to restore cytoplasmic levels of p52 subunit completely. To address this contingent, immunoblots were used to measure nuclear levels of NF-κB. The data depicted in Figure 4 show that p50, p52, and p65 are present in the nuclear fraction, and levels of the latter protein are elevated in the presence of RANKL (lanes 2–5). IL-4 incompletely blocks the RANKL-mediated nuclear increase of p52 and p65 levels (Figure 4, lanes 8–10) and has no effect on nuclear levels of p50 subunit.
Figure 4.
Residual levels of NF-κB subunits are present in nuclei under basal and IL-4–treated conditions. Nuclear extracts from cells used in Figure 3 were mixed with sample buffer and assayed for NF-κB by immunoblots.
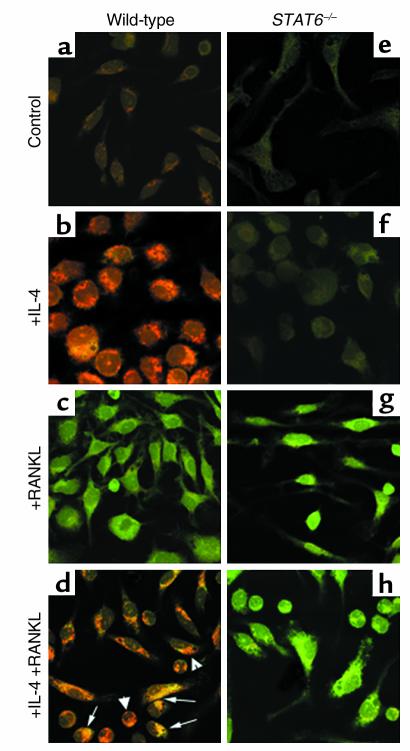
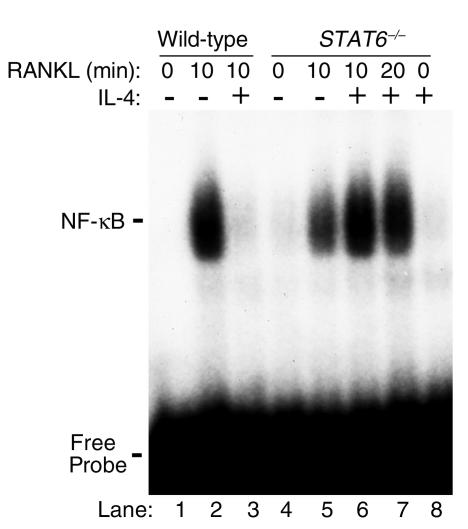
STAT6 is required for IL-4 inhibition of NF-κB activation.
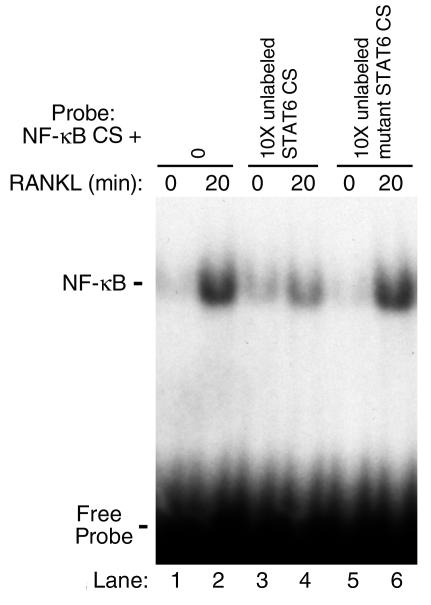
The data show that IL-4 treatment partially inhibits DNA-binding activity (Figure 2) and that the cytokine abrogates osteoclastogenesis entirely (Figure 1). Paradoxically, however, NF-κB presence in the nuclei (Figure 4), under these complete antiosteoclastic conditions (Figure 1), suggests that another mechanism, most likely a nuclear one, accounts for IL-4 attenuation of NF-κB activation and osteoclastogenesis. This also leads to the hypothesis that NF-κB subunits detected in the nuclear fraction despite IL-4 treatment must not be active. Thus it was reasoned that while some NF-κB subunits may still translocate to the nucleus in the presence of IL-4 (Figure 3), the cytokine may be preventing NF-κB DNA binding to its response elements. To address this contingent, the possible role of IL-4–induced factors in preventing DNA binding activity of NF-κB was investigated. STAT6 is an IL-4–induced transcription factor, and it was reasoned that this factor might block NF-κB activation by binding it directly and/or by preventing its DNA binding activity. To address this issue, coimmunoprecipitation and immunolocalization by confocal microscopy of NF-κB and STAT6 were performed. Immunoprecipitations failed to document direct interaction between NF-κB and STAT6 (data not shown). In contrast, however, confocal microscopy indicates that the two transcription factors indeed colocalize under IL-4– and RANKL-treated conditions (Figure 5d). This colocalization is evident by the yellow resultant color of merging red (STAT6) and green (NF-κB) colors (Figure 5d, arrows). Partial NF-κB cytoplasmic retention by STAT6 was further supported by the finding that nuclei of certain cells are dominated by STAT6 with low levels of NF-κB (Figure 5d, arrowheads). On the other hand, interaction of these two proteins was undetectable in basal conditions in various cell compartments (data not shown). Next, possible inhibition of NF-κB DNA binding activity by STAT6 was examined. The requirement of STAT6 for IL-4 inhibition of RANKL-induced NF-κB DNA binding activity was first demonstrated by EMSA using nuclear extracts prepared from STAT6 knockout cells treated with IL-4 and RANKL. The results illustrated in Figure 6 indicate that while IL-4 inhibits RANKL-induced NF-κB in wild-type cells (lane 3), the cytokine fails to block NF-κB DNA binding activity in the absence of endogenous STAT6 (lanes 6 and 7). The reason for the apparent increased NF-κB binding in IL-4–treated STAT6 knockout nuclear extracts (Figure 6, lanes 6 and 7) compared with RANKL-treated conditions (lane 5) is not clear. Although highly speculative, these differences may be due to the lack of endogenous repression otherwise exerted by STAT6. To examine further the mechanism by which STAT6 blocks NF-κB transactivation, EMSA was performed using labeled NF-κB oligonucleotide probe in the absence or presence of excess unlabeled STAT6 consensus sequence oligonucleotide or its mutated form. The results shown in Figure 7 indicate that consensus sequence STAT6 oligonucleotide inhibits NF-κB DNA binding activity (lane 4), whereas its mutated form fails to do so (lane 6). These data confirm that STAT6 acts as an antagonist perhaps allosterically or by binding to DNA sequences overlapping that of NF-κB.
Figure 5.
IL-4 and RANKL induce endogenous STAT6 and NF-κB interaction. Macrophages were grown in tissue culture coverslips for 3 days. Cells were then treated with RANKL (20 minutes) and/or IL-4 (60 minutes) or left untreated, followed by fixation with 0.25% glutaraldehyde. Cells were immunostained with STAT6 (red) and p52 NF-κB (green) antibodies alone or in combination as shown. Colocalization (d, yellow) is indicated.
Figure 6.
STAT6 is required for IL-4 inhibition of NF-κB activation. Nuclear extracts were prepared from wild-type (BALB/c) or STAT6 knockout mice treated with IL-4, with or without RANKL. NF-κB EMSA was performed as described previously.
Figure 7.
STAT6 consensus sequence inhibits NF-κB DNA-binding activity. Nuclear extracts were prepared from control and RANKL-stimulated cells as described. EMSA was carried out with 32P-ATP–labeled NF-κB oligonucleotide in the absence or presence of excess 10-fold (10×) unlabeled consensus sequence (CS) or point mutated STAT6 oligonucleotide. Specificity of the NF-κB bands is documented in Figure 2c.
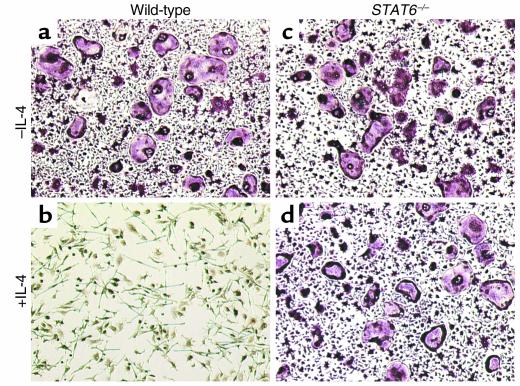
STAT6 is required for IL-4 inhibition of osteoclastogenesis.
To support further the likely role for STAT6 in IL-4 inhibition of osteoclastogenesis, the effect of IL-4 on osteoclastic differentiation of cells derived from STAT6 knockout mice was tested. The data show that IL-4 fails to block osteoclastogenesis in mice lacking STAT6 compared with their wild-type counterparts (Figure 8; compare b with d). This ex vivo evidence together with specific inhibition of NF-κB by STAT6 supports the notion that IL-4 blockade of osteoclastogenesis is mediated via STAT6 inhibition of NF-κB activation.
Figure 8.
Endogenous STAT6 is required for IL-4 inhibition of osteoclastogenesis in vitro. Osteoclast precursors harvested from STAT6 knockout mice were plated as described for Figure 1. Cells were left untreated or treated with RANKL in the absence or presence of IL-4. Osteoclast cultures were then fixed and stained for TRAP activity. TRAP+ MNC cells were counted, and the average of five independent experiments was recorded as: Control = 0, IL-4 = 0, RANKL = 210 ± 28, IL-4 + RANKL = 231 ± 17 with no statistical difference between the last two groups.
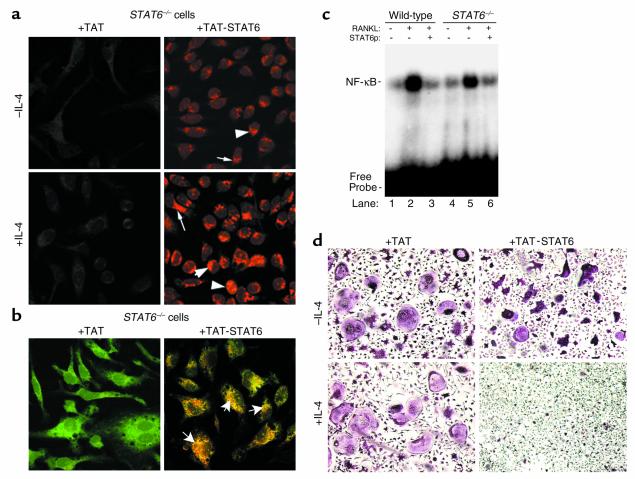
Exogenously administered STAT6 protein inhibits NF-κB DNA binding activity and osteoclastogenesis in STAT6 knockout cells.
Thus far the data indicate that STAT6 is absolutely required for IL-4 inhibition of osteoclastogenesis and deletion of the transcription factor renders this process insensitive to IL-4 action. Therefore, it was reasoned that introduction of active STAT6 to cells may be sufficient to block osteoclastogenesis. Toward this end, STAT6 was expressed and purified as a TAT-fusion protein. This system enables delivery of unlimited quantities of the functional protein into target cells in a dose-dependent fashion. By immunostaining with anti-STAT6 antibody, it was demonstrated that STAT6 is present in lysates (data not shown) and in the cytosol and nuclei of STAT6 knockout cells (Figure 9a, upper right panel arrows and arrowheads) compared with TAT-treated control cells. Furthermore, exposure of cells to IL-4 increases levels of nuclear STAT6 (Figure 9a, lower right panel, arrowheads). Next, TAT-STAT6 colocalization with NF-κB in STAT6 knockout cells treated with RANKL and IL-4 was documented (Figure 9b, arrows). Colocalization was lesser in the absence of IL-4 (data not shown) indicating that complete and potent STAT6 action requires presence of the cytokine. Furthermore, the possibility that TAT-STAT6 protein can inhibit in vitro NF-κB DNA-binding activity was tested. Nuclear extracts from wild-type and STAT6 knockout cells with or without RANKL were subjected to EMSA in the presence of TAT (control) or TAT-STAT6 protein. The data clearly indicate that TAT-STAT6 is capable of inhibiting NF-κB binding to DNA (Figure 9c, lanes 3 and 6). The next question to be addressed was whether, similar to IL-4 treatment, direct administration of STAT6 blocks osteoclastogenesis. Our results show that administration of purified TAT-STAT6 protein inhibits osteoclastogenesis in STAT6 knockout cells. Unlike NF-κB, inhibition of osteoclastogenesis, although slim in the presence of STAT6 alone (Figure 9d, upper right panel), was stronger in the presence of IL-4 (Figure 9d, lower right panel). These differences are most likely due to the fact that STAT6 is added directly to nuclear extract reaction in the former case, whereas in living cells full nuclear translocation of STAT6 requires the activation of cellular machinery by the cytokine. In any case, the data indicate that introduction of active STAT6 is sufficient to inhibit NF-κB activation and block osteoclastogenesis.
Figure 9.
Exogenously added STAT6 blocks NF-κB activation and osteoclastogenesis. (a) STAT6 knockout cells were incubated with 100 nM TAT-STAT6 for 1 hour in the absence or presence of 10 ng/ml IL-4. Cells were fixed and immunostained using anti-STAT6 antibody. (b) STAT6 knockout cells were incubated with TAT or TAT-STAT6 in the presence of IL-4 and RANKL. Cells were then fixed and stained with STAT6 and NF-κB antibodies. (c) EMSA for NF-κB was performed using control and RANKL-treated wild-type and STAT6 nuclear extracts in the presence of 2 μl TAT (lanes 2 and 5) or TAT-STAT6 (lanes 3 and 6) purified proteins (STAT6p). (d) STAT6 knockout cells were cultured with 100 nM TAT-STAT6 and RANKL in the absence or presence of 10 ng/ml IL-4 as described for Figure 1. Osteoclast cultures were fixed and TRAP-stained on day 4 of culture. TRAP+ MNC (more than three nuclei) cells were counted, and the average of triplicate wells from four independent experiments was recorded as follows: upper left panel, 226 ± 21; lower left panel, 247 ± 29; upper right panel, 186 ± 18 (P < 0.005); and lower right panel, 25 ± 11 (P < 0.0001). Note that cells treated with TAT-STAT6 appeared condensed and generally smaller, and fewer by approximately 30%, compared with controls.
To further validate the effect of TAT-STAT6, the question of whether the protein corrects failure of IL-4 to inhibit LPS-induced TNF production by STAT6-null cells (55) was examined. Indeed, in the presence of TAT-STAT6, IL-4 inhibition of LPS-induced TNF production by STAT6-null cells (57%) was shown to exceed its inhibitory effect on wild-type cells (48%) (data not shown).
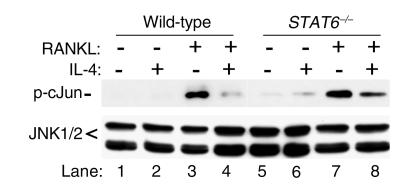
IL-4 inhibits RANKL-induced JNK activation.
We have shown recently that RANKL also activates the c-Jun N-terminal kinase (JNK) pathway (15). This MAP kinase pathway plays an important role in osteoclastogenesis, so the question of whether IL-4 inhibits RANKL-activation of JNK/cJun pathway was addressed. Osteoclast precursor cells were treated with RANKL for 20 minutes in the absence or presence (1-hour pretreatment) of IL-4. In vitro kinase assay was then performed to assess JNK activity, using its natural substrate c-Jun. The results depicted in Figure 10 indicate that although total cellular levels of JNK are unchanged, activation of JNK was abolished by IL-4. More importantly, this inhibition is partially STAT6-dependent, as IL-4 only partially blocks RANKL-activation of JNK in STAT6-null cells (Figure 10, lane 8). Although partial, this inhibition was significant (76%, n = 3). This finding indicates that although STAT6 seems to play a major role in IL-4 inhibition of RANKL-induced JNK activation, other IL-4 induced genes might also be involved.
Figure 10.
IL-4 inhibits RANKL-induced JNK activation. Wild-type and STAT6-null osteoclast precursor cells were treated for 20 minutes with RANKL (20 ng/ml) in the absence or presence (1 hour) of 10 ng/ml IL-4. C-Jun N-terminal kinase (JNK) assay was performed using c-Jun as substrate (cell signaling). Equal amounts of protein from the initial cell lysates were analyzed for total JNK protein content.
Discussion
Inflammatory processes such as in periodontitis, rheumatoid arthritis, and osteoarthritis involve massive infiltration of neutrophils, mast cells, monocytes, and macrophages to the site of inflammation (33, 56). Secreted products of these activated cells include RANKL, TNF, IL-1, and other chemokines, some of which have been shown as potent inducers of osteoclast differentiation and strongly recruit mature osteoclasts to the inflammation site and activate them (6, 11, 12, 57, 58).
Two subsets of T lymphocytes, namely Th1 and Th2 cells, have been shown to produce pro and anti-inflammatory cytokines, respectively. Proinflammatory cytokines secreted by the former cell type upon activation include RANKL, TNF, and IFN-γ. Conversely, Th2-secreted cytokines which include IL-4, IL-10, and IL-13 are immunosuppressive in nature and exert anti-inflammatory and anti-osteoclastogenic effects (33, 56, 59). The anti-inflammatory properties of IL-4 have been widely investigated and the cytokine is believed to suppress macrophage-related functions (60–64). The cytokine is present in most inflammatory sites and plays a major role as an endogenous immunoregulator (33, 35, 37, 43). Although the anti-osteoclastic properties of IL-4 have been documented, the molecular steps governing this suppression remain unknown. Therefore it was reasoned that identification of the IL-4 target cell and the molecular steps of its anti-osteoclastogenic action would provide the foundation for better therapeutic intervention.
The transcriptional regulation of many early response genes, including TNF, IL-1, and likely RANKL is largely controlled by NF-κB. Previous reports have shown that IL-4 is capable of inhibiting cytokine-induced NF-κB activation (34, 62, 65), a transcription factor essential for osteoclastogenesis (16). RANKL induction of osteoclast differentiation relies, at least in part, on NF-κB activation. Thus, it is reasonable to hypothesize that IL-4 curtails osteoclastogenesis via blockade of NF-κB activation.
The ability of IL-4 to suppress osteoclastogenesis has been demonstrated in vitro and in vivo. Given that T-cell–secreted cytokines may impact osteoclasts directly or indirectly (11, 12, 66), identification of IL-4 target cell is essential for investigating the cytokine’s mechanism of action. This study provides evidence that IL-4 directly targets osteoclast precursor cells and abrogates RANK-mediated osteoclastogenesis via inhibition of NF-κB activation by nuclear and cellular feedback mechanisms. According to this finding, it is postulated that IL-4 activates STAT6, which acts as an NF-κB inhibitor and transcriptional suppressor. Transcriptional inhibition of NF-κB and gene products regulated by NF-κB leads, presumably by feedback loop mechanism, to inhibition of kinase activation pathways leading to inhibition of IκB phosphorylation and NF-κB nuclear translocation. The data indicate, however, that despite complete IL-4 inhibition of osteoclastogenesis that of NF-κB nuclear translocation is only partial. In this regard, certain NF-κB subunits are present in the nuclear compartment under naive conditions and despite IL-4 presence. Because residual NF-κB presence and activity are occasionally detected in resting cells, two scenarios are possible. First, the finding that NF-κB is present in the nuclear compartment in the face of IL-4 may represent partial failure of the cytokine to prevent cytosolic to nuclear translocation. Second, basal levels of certain NF-κB subunits are constantly present in the nucleus to maintain basic cellular functions not related to osteoclastogenesis. In any case, it is clear that IL-4 potently abrogates osteoclastogenesis despite the nuclear presence of these NF-κB subunits. Although it is tempting to hypothesize that IL-4 inhibition of osteoclastogenesis may be independent of NF-κB, the current evidence (Figures 6 and 7) points out that an intranuclear mechanism(s) may account for NF-κB inactivation and blockade of osteoclastogenesis. These observations are best supported by the current findings that IL-4 induces activation of STAT6, which acts as an antagonist preventing NF-κB DNA binding activity. These findings are consistent with those of a previous study in which IL-4 was found to inhibit TNF-induced NF-κB DNA binding due to STAT6 antagonism in the E-selectin gene promoter of endothelial cells (65). Support for IL-4 nuclear inhibition of NF-κB is also provided by data in the present study showing endogenous interaction of STAT6 and NF-κB after IL-4 and RANKL treatment (Figure 5). I favor interaction of the two transcription factors as documented by confocal microscopy and speculate that failure of coimmunoprecipitating STAT6 and NF-κB is the result of one or a combination of the following reasons. First, confocal microscopy directly reflects endogenous events, as cells undergo fixation before any further manipulations. Second, it is likely that interaction of these two proteins requires binding to DNA, as occurs in vivo. Lysis of cells may affect these complexed interactions. Third, interactions between transcription factors and DNA are naturally weak and subject to dissociation in detergent-based lysis buffers. The data raise the possibility that STAT6 may be the principal player in IL-4 inhibition of osteoclastogenesis. In this regard, given that NF-κB regulates a large number of genes including its own, it is conceivable that STAT6 inhibition of NF-κB activation leads, in a negative feedback loop, to inhibition of cellular levels and activation of IKKs, IκB, and NF-κB. This scenario may also help explain the partial and redundant inhibition of cellular levels of NF-κB, a gene that regulates not only osteoclastogenesis but numerous other cellular functions.
Direct inhibition of NF-κB activation and osteoclastogenesis by STAT6 is best demonstrated by the findings showing that introduction of STAT6 protein inhibits NF-κB DNA binding activity and blocks osteoclastogenesis. While inhibition of NF-κB binding to DNA seems readily attainable with TAT-STAT6 (Figure 9c), potent inhibition of osteoclastogenesis with this protein requires additional activation with IL-4 (Figure 9d). These differences are most likely due to the fact that STAT6 is added directly to nuclear extract reaction in the former case, whereas in living cells full nuclear translocation of STAT6 requires the activation of cellular machinery by the cytokine. This method of direct protein introduction into cells, in a nonspecific manner and without the aid of chemical agents, presents itself as promising delivery system in vitro and in vivo (51–53, 67).
We have shown recently that activation of multiple pathways, including NF-κB and c-Jun, mediates RANKL induction of osteoclastogenesis (15). Given the redundancy of genes affecting osteoclast development, it is conceivable that RANKL-induced genes, other than NF-κB, may be regulated by IL-4. One such candidate is the c-Jun/AP-1 pathway. Indeed, this study finds that IL-4 treatment of osteoclast precursors significantly inhibits c-Jun phosphorylation by its upstream c-Jun N-terminal kinase, JNK (Figure 10). It further demonstrates that this inhibition is a partially STAT6-dependent event (76%; compare lanes 7 and 8). This novel finding indicates that inhibition of RANKL-induced JNK by IL-4 may require not only STAT6 but also activation of other genes.
The osteoclastogenic factors RANKL and its decoy receptor osteoprotegerin (OPG) are secreted by osteoblast/stromal cells and tightly regulate osteoclast differentiation (8, 68). Regulation of OPG by IL-4, if any, in stromal cells is currently unknown. To exclude the possibility that IL-4 inhibition of osteoclastogenesis is due to enhancing OPG secretion by contaminating stromal cells, a nearly 98% pure population of monocytes/macrophages was obtained using magnetic bead-conjugated CD11b antibody purification method. In this nearly pure population of marrow macrophages, OPG was not detected and IL-4 abrogated osteoclastogenesis. Thus, complete IL-4 inhibition of osteoclastogenesis in marrow macrophages appears direct and independent of OPG.
IL-4 ligation to its receptor prompts activation of various molecules, most notably the lipid, phospho-inositol-3-kinase (PI3K), and the transcriptional activator, STAT6 (34, 69, 70). Wortmannin is a potent inhibitor of PI3K (70); however, under this study’s experimental conditions, the IL-4–osteoclast inhibitory effect was irreversible. Thus, I believe IL-4 inhibition of osteoclastogenesis may be independent of PI3K.
Activation and translocation of STAT6 by IL-4 occur rapidly. Further observations made in this study indicate that a short pre-exposure to IL-4 (30 minutes) or simultaneous exposure of cells to IL-4 and RANKL was sufficient to completely block NF-κB activation and osteoclastogenesis. In this regard, an initial, single exposure of osteoclast progenitor cells to IL-4 activates signals sufficient to attenuate osteoclastogenesis despite subsequent multiple additions of RANKL. The most compelling evidence for a key role for STAT6-mediated IL-4 inhibition of osteoclastogenesis lies in the novel finding that the cytokine failed to abrogate osteoclastogenesis in mice lacking the STAT6 gene, a function that was restored by in vitro introduction of TAT-STAT6 fusion protein.
In summary, this study delineates the molecular mechanism of IL-4 inhibition of osteoclastogenesis. The findings of this study point out that IL-4 inhibits NF-κB nuclear translocation and activation by RANKL, in STAT6-dependent manner. IL-4 also inhibits RANKL-activation of the MAP kinase pathway, JNK-cJun, at least in part by a similar mechanism. These events lead to attenuation of RANKL-induced osteoclastogenesis. The data strongly suggest that STAT6 may be acting as a transcriptional inhibitor that inhibits NF-κB target genes, including NF-κB itself. These findings are the first to clearly identify molecular steps of IL-4 inhibition of RANKL-mediated osteoclastogenesis, thus providing clear therapeutic targets in cases of excessive bone loss diseases due to increased osteoclastic activity.
Acknowledgments
This work was supported in part by an Arthritis Foundation Award, a Monsanto-Searle grant, and by a grant from the NIH (DE 13754). The author thanks Aditi Mukherjee and Yan-Hong Zhang for technical assistant, Steven Dowdy for assistance with TAT-protein transduction system, and Kenneth Murphy for providing the STAT6 cDNA and for helpful discussions.
References
- 1.Teitelbaum SL, Abu-Amer Y, Ross FP. Molecular mechanisms of bone resorption. J Cell Biochem. 1995;59:1–10. doi: 10.1002/jcb.240590102. [DOI] [PubMed] [Google Scholar]
- 2.Teitelbaum SL, Tondravi MM, Ross FP. Osteoclasts, macrophages, and the molecular mechanisms of bone resorption. J Leukoc Biol. 1997;61:381–388. doi: 10.1002/jlb.61.4.381. [DOI] [PubMed] [Google Scholar]
- 3.Udagawa N, et al. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc Natl Acad Sci USA. 1990;87:7260–7264. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi N, et al. Osteoclast-like cell formation and its regulation by osteotropic hormones in mouse bone marrow cultures. Endocrinology. 1988;122:1373–1382. doi: 10.1210/endo-122-4-1373. [DOI] [PubMed] [Google Scholar]
- 5.Udagawa N, et al. The bone marrow-derived stromal cell lines MC3T3-G2/PA6 and ST2 support osteoclast-like cell differentiation in cocultures with mouse spleen cells. Endocrinology. 1989;125:1805–1813. doi: 10.1210/endo-125-4-1805. [DOI] [PubMed] [Google Scholar]
- 6.Abu-Amer Y, Ross FP, Edwards J, Teitelbaum SL. Lipopolysaccharide-stimulated osteoclastogenesis is mediated by tumor necrosis factor via its p55 receptor. J Clin Invest. 1997;100:1557–1565. doi: 10.1172/JCI119679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu H, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci USA. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacey DL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 9.Suda T, Udagawa N, Nakamura I, Miyaura C, Takahashi N. Modulation of osteoclast differentiation by local factors. Bone. 1996;17:87S–91S. doi: 10.1016/8756-3282(95)00185-g. [DOI] [PubMed] [Google Scholar]
- 10.Suda T, Nakamura I, Jimi E, Takahashi N. Regulation of osteoclast function. J Bone Miner Res. 1997;12:869–879. doi: 10.1359/jbmr.1997.12.6.869. [DOI] [PubMed] [Google Scholar]
- 11.Burgess TL, et al. The ligand for osteoprotegerin (OPGL) directly activates mature osteoclasts. J Cell Biol. 1999;145:527–538. doi: 10.1083/jcb.145.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong YY, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 13.Wong BR, Lee SY, Vologodskia M, Steinman RM, Choi Y. The TRAF family of signal transducers mediates NF-kB activation by the TRANCE receptor. J Biol Chem. 1998;273:28335–28359. doi: 10.1074/jbc.273.43.28355. [DOI] [PubMed] [Google Scholar]
- 14.Darnay BG, Ni J, Moore PA, Aggarwal B. Characterization of the intracellular domain of receptor activator of NF-kB (RANK) J Biol Chem. 1998;273:20551–20555. doi: 10.1074/jbc.273.32.20551. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YH, Heulsmann A, Tondravi MM, Mukherjee A, Abu-Amer Y. Tumor necrosis factor-alpha (TNF) stimulates RANKL-induced osteoclastogenesis via coupling of TNF type 1 receptor and RANK signaling pathways. J Biol Chem. 2000;276:563–568. doi: 10.1074/jbc.M008198200. [DOI] [PubMed] [Google Scholar]
- 16.Iotsova V, et al. Osteopetrosis in mice lacking NFkB1 and NFkB2. Nat Med. 1997;3:1285–1289. doi: 10.1038/nm1197-1285. [DOI] [PubMed] [Google Scholar]
- 17.Abu-Amer Y, Tondravi MM. NFkB and bone: the breaking point. Nat Med. 1997;3:1189–1190. doi: 10.1038/nm1197-1189. [DOI] [PubMed] [Google Scholar]
- 18.Thanos D, Maniatis T. NF-kappaB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 19.Stancovski I, Baltimore D. NF-kB activation: the IkB kinase revealed? Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 20.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 21.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 22.Regnier CH, et al. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 23.Mercurio F, et al. IKK1 and IKK2: cytokine-activated IkB kinases essential for NF-kB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 24.Malinin NL, Boldin MP, Kovalenko AV, Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 25.Singh S, Darnay BG, Aggarwal BB. Site-specific tyrosine phosphorylation of IkBalpha negatively regulates its inducible phosphorylation and degradation. J Biol Chem. 1996;271:31049–31054. doi: 10.1074/jbc.271.49.31049. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Amer Y, et al. Tumor necrosis factor-alpha activation of nuclear transcription factor-kappaB in marrow macrophages is mediated by c-Src tyrosine phosphorylation of Ikappa Balpha. J Biol Chem. 1998;273:29417–29423. doi: 10.1074/jbc.273.45.29417. [DOI] [PubMed] [Google Scholar]
- 27.Lacey DL, et al. Interleukin-4, interferon gamma and prostaglandin E impact the osteoclast forming potential of murine bone marrow macrophages in vitro. Endocrinology. 1995;136:2367–2376. doi: 10.1210/endo.136.6.7750457. [DOI] [PubMed] [Google Scholar]
- 28.Mundy GR. Cytokine and bone remodeling. Osteoporosis. 1996;11:302–310. [Google Scholar]
- 29.Okada Y, et al. Short term treatment of recombinant IL-4 rapidly inhibits bone formation in normal and ovariectomized mice. Bone. 1998;22:361–365. doi: 10.1016/s8756-3282(97)00296-2. [DOI] [PubMed] [Google Scholar]
- 30.Mundy GR. Role of cytokines in bone resorption. J Cell Biochem. 1993;53:296–300. doi: 10.1002/jcb.240530405. [DOI] [PubMed] [Google Scholar]
- 31.Bizzarri C, et al. Interleukin-4 inhibits bone resorption and acutely increases cytosolic Ca2+ in murine osteoclasts. J Biol Chem. 1994;269:13817–13824. [PubMed] [Google Scholar]
- 32.Duff GW. Cytokines and acute phase proteins in rheumatoid arthritis. Scand J Rheumatol. 1994;100:9–19. doi: 10.3109/03009749409095197. [DOI] [PubMed] [Google Scholar]
- 33.Allen JE, Maizels RM. Th1-Th2: reliable paradigm or dangerous dogma? Immunol Today. 1997;18:387–392. doi: 10.1016/s0167-5699(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 34.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 35.Wagner S, Fritz P, Einsele H, Sell S, Saal JG. Evaluation of synovial cytokine patterns in rheumatoid arthritis and osteoarthritis by quantitative reverse transcription polymerase chain reaction. Rheumatol Int. 1997;16:191–196. doi: 10.1007/BF01330295. [DOI] [PubMed] [Google Scholar]
- 36.Ulfgren AK, Lindblad S, Klareskog L, Andersson J, Andersson U. Detection of cytokine producing cells in the synovial membrane from patients with rheumatoid arthritis. Ann Rheum Dis. 1995;54:654–661. doi: 10.1136/ard.54.8.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlaak JF, Pfers I, Meyer-Zum KH, Marker-Hermann E. Different cytokine profiles in the synovial fluid of patients with osteoarthritis, rheumatoid arthritis and seronegative spondylarthropathies. Clin Exp Rheumatol. 1996;14:155–162. [PubMed] [Google Scholar]
- 38.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 39.Oakes SA, et al. Signaling via IL-2 and IL-4 in JAK3-deficient severe combined immunodeficiency lymphocytes: JAK3-dependent and independent pathways. Immunity. 1996;5:605–615. doi: 10.1016/s1074-7613(00)80274-5. [DOI] [PubMed] [Google Scholar]
- 40.Rolling C, Treton D, Pellegrini S, Galanaud P, Richard Y. IL4 and IL13 receptors share the gamma c chain and activate STAT6, STAT3 and STAT5 proteins in normal human B cells. FEBS Lett. 1996;393:53–56. doi: 10.1016/0014-5793(96)00835-6. [DOI] [PubMed] [Google Scholar]
- 41.Ohmori Y, Hamilton TA. STAT6 is required for the anti-inflammatory activity of interleukin-4 in mouse peritoneal macrophages. J Biol Chem. 1998;273:29202–29209. doi: 10.1074/jbc.273.44.29202. [DOI] [PubMed] [Google Scholar]
- 42.Takeda K, Kishimoto T, Akira S. STAT6: its role in interleukin 4-mediated biological functions. J Mol Med. 1997;75:317–326. doi: 10.1007/s001090050117. [DOI] [PubMed] [Google Scholar]
- 43.Miossec P, et al. Interleukin-4 inhibits bone resorption through an effect on osteoclasts and proinflammatory cytokines in an ex-vivo model of bone resorption in RA. Arthritis Rheum. 1994;37:1715–1722. doi: 10.1002/art.1780371202. [DOI] [PubMed] [Google Scholar]
- 44.Riancho JA, Zarrabeitia MT, Mundy GR, Yoneda T, Gonzalez-Macias J. Effects of interleukin-4 on the formation of macrophages and osteoclast-like cells. J Bone Miner Res. 1993;8:1337–1344. doi: 10.1002/jbmr.5650081108. [DOI] [PubMed] [Google Scholar]
- 45.Riancho JA, Zarrabeitia MT, Gonzalez-Macias J. Interleukin-4 modulates osteoclast differentiation and inhibits the formation of resorption pits in mouse osteoclast cultures. Biochem Biophys Res Commun. 1993;196:678–685. doi: 10.1006/bbrc.1993.2303. [DOI] [PubMed] [Google Scholar]
- 46.Abu-Amer Y, Erdmann J, Alexopoulou L, Kollias G, Teitelbaum SL. Tumor necrosis factor receptors types 1 and 2 differentially regulate osteoclastogenesis. J Biol Chem. 2000;275:27307–27310. doi: 10.1074/jbc.M003886200. [DOI] [PubMed] [Google Scholar]
- 47.Clohisy DR, Chappel JC, Teitelbaum SL. Bone marrow-derived mononuclear phagocytes autoregulate mannose receptor expression. J Biol Chem. 1989;264:5370–5377. [PubMed] [Google Scholar]
- 48.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Tetsuka T, Srivastava SK, Morrison AR. Tyrosine kinase inhibitors, genistein and herbimycin A, do not block interleukin-1 beta-induced activation of NF-kappa B in rat mesangial cells. Biochem Biophys Res Commun. 1996;218:808–812. doi: 10.1006/bbrc.1996.0144. [DOI] [PubMed] [Google Scholar]
- 50.Drouet C, Shakhov AN, Jongeneel CV. Enhancers and transcription factors controlling the inducibility of the tumor necrosis factor-alpha promoter in primary macrophages. J Immunol. 1991;147:1694–1700. [PubMed] [Google Scholar]
- 51.Nagahara H, et al. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 52.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 53.Schwarze SR, Hruska KA, Dowdy SF. Protein transduction: unrestricted delivery into all cells? Trends Cell Biol. 2000;10:290–295. doi: 10.1016/s0962-8924(00)01771-2. [DOI] [PubMed] [Google Scholar]
- 54.Kitazawa S, Ross FP, McHugh K, Teitelbaum SL. Interleukin-4 induces expression of the integrin αvβ3 via transactivation of the β3 gene. J Biol Chem. 1995;270:4115–4120. doi: 10.1074/jbc.270.8.4115. [DOI] [PubMed] [Google Scholar]
- 55.Levings MK, Schrader JW. IL-4 inhibits the production of TNF-alpha and IL-12 by STAT6-dependent and -independent mechanisms. J Immunol. 1999;162:5224–5229. [PubMed] [Google Scholar]
- 56.Rocken M, Racke M, Shevach EM. IL-4-induced immune deviation as antigen-specific therapy for inflammatory autoimmune disease. Immunol Today. 1996;17:225–231. doi: 10.1016/0167-5699(96)80556-1. [DOI] [PubMed] [Google Scholar]
- 57.Pfeilschifter J, Chenu C, Bird A, Mundy GR, Roodman GD. Interleukin-1 and tumor necrosis factor stimulate the formation of human osteoclast-like cells in vitro. J Bone Miner Res. 1989;4:113–118. doi: 10.1002/jbmr.5650040116. [DOI] [PubMed] [Google Scholar]
- 58.Jimi E, et al. Interleukin 1 induces multinucleation and bone-resorbing activity of osteoclasts in the absence of osteoblasts/stromal cells. Exp Cell Res. 1999;247:84–93. doi: 10.1006/excr.1998.4320. [DOI] [PubMed] [Google Scholar]
- 59.Kong YY, et al. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature. 1999;402:304–309. doi: 10.1038/46303. [DOI] [PubMed] [Google Scholar]
- 60.McNally AK, DeFife KM, Anderson JM. Interleukin-4-induced macrophage fusion is prevented by inhibitors of mannose receptor activity. Am J Pathol. 1996;149:975–985. [PMC free article] [PubMed] [Google Scholar]
- 61.McNally AK, Anderson JM. Interleukin-4 induces foreign body giant cells from human monocytes/macrophages. Differential lymphokine regulation of macrophage fusion leads to morphological variants of multinucleated giant cells. Am J Pathol. 1995;147:1487–1499. [PMC free article] [PubMed] [Google Scholar]
- 62.Manna SK, Aggarwal BB. Interleukin-4 down-regulates both forms of tumor necrosis factor receptor and receptor-mediated apoptosis, NF-kappaB, AP-1, and c-Jun N-terminal kinase. Comparison with interleukin-13. J Biol Chem. 1998;273:33333–33341. doi: 10.1074/jbc.273.50.33333. [DOI] [PubMed] [Google Scholar]
- 63.Kazazi F, Chang J, Lopez A, Vadas M, Cunningham AL. Interleukin 4 and human immunodeficiency virus stimulate LFA-1-ICAM-1-mediated aggregation of monocytes and subsequent giant cell formation. J Gen Virol. 1994;75:2795–2802. doi: 10.1099/0022-1317-75-10-2795. [DOI] [PubMed] [Google Scholar]
- 64.Andersson A, Grunewald SM, Duschl A, Fischer A, DiSanto JP. Mouse macrophage development in the absence of the common gamma chain: defining receptor complexes responsible for IL-4 and IL-13 signaling. Eur J Immunol. 1997;27:1762–1768. doi: 10.1002/eji.1830270725. [DOI] [PubMed] [Google Scholar]
- 65.Bennett BL, Cruz R, Lacson RG, Manning AM. Interleukin-4 suppression of tumor necrosis factor alpha-stimulated E-selectin gene transcription is mediated by STAT6 antagonism of NF-kappaB. J Biol Chem. 1997;272:10212–10219. doi: 10.1074/jbc.272.15.10212. [DOI] [PubMed] [Google Scholar]
- 66.Kotake S, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 2000;103:1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abu-Amer Y, Dowdy SF, Ross F, Teitelbaum SL. TAT fusion proteins containing dominant-negative IkB block osteoclastogenesis. Bone. 1999;13(Suppl.):S112. doi: 10.1074/jbc.M104725200. (Abstr.) [DOI] [PubMed] [Google Scholar]
- 68.Simonet WS, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 69.Onoe AP, et al. IL-13 and IL-4 inhibit bone resorption by suppressing COX-2-dependent prostaglandin synthesis in osteoblasts. J Immunol. 1996;156:758–764. [PubMed] [Google Scholar]
- 70.Bracke M, Coffer PJ, Lammers JW, Koenderman L. Analysis of signal transduction pathways regulating cytokine-mediated Fc receptor activation on human eosinophils. J Immunol. 1998;161:6768–6774. [PubMed] [Google Scholar]