Abstract
The discovery of many fatal human disorders resulting from impaired peroxisomal protein import makes the functional characterization of human peroxins critical. As part of our attempt to identify novel human genes and gene products involved in the import of peroxisomal proteins, we raised antisera against peroxisomal membrane proteins. One such antiserum inhibited peroxisomal protein import in semipermeabilized mammalian cells. This “import inhibiting” antiserum, ab-MF3, specifically recognized a 57-kDa protein. Immunoblot analysis of rat liver subcellular fractions demonstrated that this protein was present exclusively in peroxisomal membranes. Functional analysis revealed that this 57-kDa molecule bound the PTS1 receptor, Pex5p, in ligand blots, suggesting it is a docking site on the peroxisomal membrane. Previous studies have identified two yeast proteins, Pex14p and Pex13p, as Pex5p-binding proteins. To facilitate the biochemical analysis of peroxisomal membrane docking proteins, we cloned and expressed the previously unidentified human Pex14p, as well as a human Pex13p that is 39 aa longer than previously reported. Recombinant Pex14p was specifically recognized by the “import inhibiting” ab-MF3 and bound Pex5p and the Src homology 3 (SH3) domain of Pex13p in ligand blots. These studies demonstrate that the ab-MF3-immunoreactive, 57-kDa peroxisomal membrane protein is Pex14p. Furthermore, this peroxin interacts with Pex5p and Pex13p(SH3) and is directly required for peroxisomal protein import.
The existence of a variety of debilitating and often lethal disorders associated with functionally defective peroxisomes underscores the importance of these organelles in human cellular physiology. In recent years, a dramatic increase in our understanding of peroxisomal protein import and the biogenesis of this organelle has been realized. Through genetic and biochemical approaches in a variety of eukaryotes, 18 proteins, called peroxins (1), have been implicated in peroxisome assembly/import (2–4). Although the functions of many of these peroxins awaits elucidation, especially in humans, certain aspects of the import process are well understood. For example, thoroughly analyzed are the peroxisomal targeting signals (PTSs) for matrix proteins and the receptors that bind them (5).
With only a few exceptions, peroxisomal matrix proteins contain the carboxyl-terminal amino acids Ser-Lys-Leu or a closely related sequence. These sequences constitute the PTS1 (6). A second sequence, PTS2, is an amino-terminal stretch of nine amino acids (7, 8) and thus far is found in only a select few proteins. Proteinaceous receptors that specifically recognize these PTSs and direct them to the peroxisome membrane also have been described (4).
PTS1 is recognized by the tetratricopeptide repeat family protein, Pex5p, whereas PTS2 is recognized by the WD40 protein, Pex7p. Cells lacking functional Pex5p are unable to import PTS1-containing proteins but, in most cases, are unaffected in their capacity to import PTS2-containing substrates. The converse is also true; PTS2—but not PTS1—protein import is abolished in cells lacking Pex7p. Although these results suggest import pathways with distinct machineries for at least the early stages of import, recent evidence points to a “shared” translocation apparatus (9).
Because peroxisomal protein import is posttranslational (10), recognition of the PTS1 by its particular receptor presumably occurs in the cytosol or at the peroxisome membrane. The precise mechanism by which these receptor–ligand complexes are then directed to downstream elements of the peroxisomal membrane transport apparatus and what dissociates them either there or within the organelle are only beginning to be understood. Insight into this process has been provided by the identification of “docking proteins,” molecules localized to the peroxisome membrane that bind the PTS receptors.
Pex13p, identified in the yeasts Saccharomyces cerevisiae (11, 12) and Pichia pastoris (13), as well as in human cells (13), is a Src homology 3 (SH3) domain-containing peroxisomal integral membrane protein that binds Pex5p. Its interaction with the PTS1 receptor is mediated by the SH3 domain of Pex13p, and in a pex13Δ mutant, PTS1-protein import is eliminated. Interestingly, PTS2-protein import is also abolished in these cells, although the basis for this is unclear. A second docking protein, identified in the yeasts S. cerevisiae (9, 14) and Hansenula polymorpha (15), is the tightly associated peroxisomal membrane protein, Pex14p. This critical protein interacts with both the PTS1 and PTS2 receptors, as well as with Pex13p and the recently described peroxin, Pex17p (2). Importantly, loss of functional Pex14p results in defects in both of the PTS-dependent import pathways. The precise functions and regulation of this complex are not known, nor is it clear whether or not a similar complex of multifunctional peroxins exists in mammalian cells.
Our work to identify components of the mammalian peroxisomal protein import machinery involves the development of high-affinity antibodies to specific peroxisomal membrane proteins. One antiserum specifically reacts with a rat liver 57-kDa peroxisomal integral membrane protein that, importantly, also is recognized by (human) Pex5p in a ligand blot-binding assay. We show here that this protein is Pex14p, and, using a biochemically defined in vitro import system, we directly implicate Pex14p in PTS1-protein import. We also report the cloning of the human homologue of this important peroxin, as well as a previously unrecognized form of human Pex13p that is 39 aa longer than the one reported earlier (13).
MATERIALS AND METHODS
Animals.
Animals were maintained as described previously (16), and animal care approval was granted by the local institutional ethics committee.
Purification and Subfractionation of Peroxisomes.
Rat liver homogenates were fractionated as described (16) in the presence of protease inhibitors. For the generation of antisera, the highly purified peroxisomes were resuspended in buffer A [10 mM Mops/1 mM EDTA/1 mM DTT/0.1% (vol/vol) ethanol, pH 8.0] and sonicated. After centrifugation for 1 h at 100,000 × g, the pellet was resuspended in buffer A and the entire procedure was repeated. The combined supernatant (S1) contained the matrix proteins (71% of total peroxisomal protein). The pellet (P1) was resuspended in buffer A supplemented with 1 M NaCl and subjected to the same procedure yielding supernatant S2 (peripheral membrane proteins, 7% of total peroxisomal protein) and pellet P2. P2 was resuspended in buffer B [10 mM pyrophosphate/1 mM EDTA/1 mM DTT/0.1% ethanol (vol/vol), pH 9.0], and after sonication and centrifugation, the supernatant S3 (peripheral membrane proteins, 0.6% of total peroxisomal protein) and pellet P3 were obtained. Finally, the P3 fraction was subjected to carbonate extraction (17). This extraction resulted in the supernatant S4 (10.4% of total peroxisomal protein) yielding peripheral membrane proteins and urate oxidase, and a pellet P4 containing the integral membrane proteins (IMPs, 11% of total peroxisomal protein). Identification on Western blots of specific peroxisomal matrix proteins was made by probing similar blots with protein-specific antibodies (18).
Generation of Polyclonal Antisera.
Ab-MF3 was raised against peroxisomal integral membrane proteins with a molecular mass between 30 and 58 kDa. One hundred micrograms of the P4-fraction was separated in an SDS/10% (wt/vol) polyacrylamide gel and transferred onto nitrocellulose. The 30- to 58-kDa region was cut out and the nitrocellulose membrane was dissolved by adding dimethyl sulfoxide as described (19). An equal amount of Freund’s complete adjuvant (priming dose) or Freund’s incomplete adjuvant (successive boosting doses) was added, and after sonication, the emulsified solution was injected subcutaneously into a rabbit (20). The antiserum MF16 (ab-MF16) was raised against peroxisomal matrix proteins. For this, 100 μg of the total S1-fraction was mixed with Freund’s (in)complete adjuvant and the antiserum was generated in the same way as for ab-MF3.
Peroxisomal Import Assay in Permeabilized CHO Cells.
The permeabilized-cell import assay was performed as described (21). To test the effect of exogenous peptides or antisera, 5 μl of these reagents was added to the import mix. To localize HSA-SKL, we used a goat anti-HSA primary antibody and fluorescein isothiocyanate-labeled rabbit anti-goat secondary IgGs. At least 250 cells were analyzed and the number with detectable peroxisomal HSA-SKL was determined. Under standard conditions, 70–80% of the cells contained such peroxisomal HSA-SKL. To compare experiments, the percentage of CHO cells with detectable peroxisomal HSA-SKL in the internal “standard condition” was defined as 100%. When peroxisomal protein import is impaired below a specific threshold, peroxisomal HSA-SKL is not detected. Partial inhibition results in a mixed population with some cells displaying HSA-SKL import, and others not. It is this change in detectable HSA-SKL import that is mirrored in the percentage of cells that exhibit import in our assay. To ensure that the results were reliable, we performed the import experiments described here by using samples coded in a blind fashion.
Ligand Blot Assay.
Peroxisomal matrix proteins were prepared by treating rat liver peroxisomes for 1 h at 4°C with (hypotonic) lysis buffer (10 mM Tris⋅HCl, pH 8.0) and centrifuging for 1 h at 100,000 × g. Pellets from the hypotonic lysis step were extracted on ice for 16 h with 100 mM sodium carbonate (pH 11.0) and again centrifuged to obtain peroxisomal membrane proteins in the pellet. These molecules were separated by SDS/PAGE and transferred to nitrocellulose membranes. The proteins were renatured for 2 h at 4°C in buffer A [50 mM Tris⋅HCl, pH 7.5/100 mM potassium acetate/150 mM NaCl/1 mM DTT/5 mM MgCl2/1 mM EDTA/0.3% (vol/vol) Tween-20/100 μM ZnCl2/5% (wt/vol) nonfat milk/100 mM methionine, as modified from Cuervo and Dice (22)]. 35S-labeled HsPex5p was prepared by in vitro transcription/translation in a rabbit reticulocyte lysate (TnT Coupled Reticulocyte Lysate Systems, Promega). [35S]HsPex5p (23) constituted greater than 95% of the 35S-labeled protein synthesized (data not shown). The membranes were then incubated overnight at 4°C with 25 ml of buffer A containing 25 μl of the in vitro transcription/translation lysate. After washing once with buffer A, bound [35S]HsPex5p was visualized with a Molecular Dynamics PhosphorImager.
cDNA Cloning.
Standard procedures were used for cDNA cloning (24) and sequencing (25). Direct cloning of DNA amplified by Pfu polymerase into the TOPO TA cloning vector (Invitrogen) was performed as described by the manufacturer. Database searches were performed by using the blast algorithm (26). To generate the plasmid encoding the biotinylated HsPex5p (pMF52), the cDNA encoding HsPex5p was transferred from pBE40 (23) into the PinPoint expression vector Xa2 (Promega).
Cloning and Expression of the Human PEX14.
By probing the human expressed sequence tag (EST) database with ScPex14p and HpPex14p, we found one EST (GenBank accession no. H16035) encoding the putative full-length human homologue of Pex14p. The cDNA encoding the putative ORF was sequenced in both directions and transferred into the pCR2.1-TOPO vector by PCR (Pfu-polymerase, primers 5′-ATGGCGTCCTCGGAGCAGGC-3′ and 5′-GGGCTGGAGGCAGCAGGC-3′), and transformants (pMF34) with a HsPex14 cDNA oriented in the opposite direction to the LacZα DNA were selected. pMF42, encoding the biotinylated HsPex14 fusion protein, was generated by transferring the 1.2-kb EcoRV/KpnI fragment of pMF34 into the PinPoint Xa2 expression vector digested with PvuII and KpnI. pMF44, encoding the (His)6-tagged HsPex14p, was generated by transferring the 1.2-kb EcoRV/HindIII fragment of pMF34 into the pQE30 vector (Qiagen) digested with SmaI and HindIII. To check the expression of the biotinylated HsPex14p, bacterial lysates fractionated by SDS/PAGE were transferred to nitrocellulose by semidry blotting, and the biotinylated fusion protein was detected with streptavidin alkaline phosphatase. The expression and purification of (His)6-HsPex14p was performed according to the manufacturer’s instructions by using Ni-NTA agarose. Ligand blotting of freshly prepared bacterial lysates (approximately 20 μg of protein) was as described for the rat liver peroxisome subfractions above.
Cloning of Human PEX13 and Expression of Its SH3 Domain.
Probing the database with HsPex13p (GenBank accession no. U71374) revealed the existence of a human EST (GenBank accession no. AA295033) that encoded, in addition to the HsPex13p sequence reported (13), an additional 39 amino-terminal amino acids. The cDNA sequence encoding this longer form of HsPex13p was confirmed by sequencing. pMF59, encoding the reported Pex5p-binding SH3-binding domain (13), was generated by transferring the 498-bp SspI/BglII fragment into the PinPoint Xa1 expression vector digested with NruI and BglII. pMF40, encoding (His)6-HsPex13p(SH3), was generated by transferring the 560-bp PstI/SphI fragment into pQE30 (digested with PstI and SphI).
RESULTS
Ab-MF3 Specifically Recognizes a 57-kDa Peroxisomal Membrane Protein.
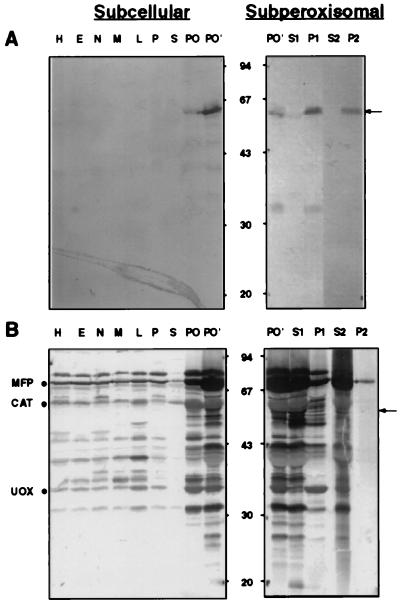
We raised a rabbit antiserum (designated hereafter as ab-MF3) against peroxisomal integral membrane proteins with molecular masses between 30 and 58 kDa. Immunoblot analysis of rat liver subcellular fractions using this antiserum demonstrated that ab-MF3 specifically bound to a 57-kDa protein (Fig. 1A). By using ab-MF16, a polyspecific antiserum recognizing more than 25 peroxisomal (matrix) proteins (Fig. 1B), we showed that the 57-kDa ab-MF3 immunoreactive protein colocalized with the peroxisomal marker enzymes urate oxidase, catalase, and the inducible multifunctional protein (Fig. 1 A and B, Subcellular). Probing peroxisomal subfractions with ab-MF3 revealed that the 57-kDa immunoreactive peroxisomal protein behaved as a tightly associated membrane protein (Fig. 1A, Subperoxisomal). This 57-kDa peroxisomal membrane protein (hereafter referred to as PMP57) was seen faintly in peroxisomes of untreated rat liver (Fig. 1A, Subcellular, PO), but was markedly induced by the hypolipidemic (peroxisome-proliferating) drug clofibrate (Fig. 1A, Subcellular, PO′). Furthermore, PMP57 was easily degraded to a 35-kDa protein (Fig. 1A, Subperoxisomal); that this immune reactive fragment is indeed a proteolytic degradation product of PMP57 is suggested by the observation that its appearance is correlated with the disappearance of the holomolecule (data not shown). Furthermore, when antibodies bound either to the 57-kDa or the 35-kDa protein were eluted, they each cross-reacted with the other protein (data not shown).
Figure 1.
Distribution of proteins cross-reacting with ab-MF3 and ab-MF16 in rat liver subcellular fractions. Protein (100 μg), present in rat liver homogenates, subcellular fractions, and purified peroxisomes, or, in the case of the peroxisomal subfractions, the amount of protein extracted from 100 μg of total peroxisomal proteins, was subjected to SDS/PAGE, transferred to nitrocellulose, and immunoblotted with ab-MF3 (A) or ab-MF16 (B). H, homogenate; E, postnuclear supernatant; N, nuclear fraction; M, heavy mitochondrial fraction; L, light mitochondrial fraction; P, microsomal fraction; S, cytosol; PO, purified total peroxisomes; and PO′, purified total peroxisomes of a clofibrate-treated rat. S1, P1, S2, and P2 are peroxisomal subfractions corresponding to the (S) supernatant and (P) pellet of (1) 10 mM pyrophosphate (pH 9.0) and (2) 100 mM sodium carbonate (pH 11.0) extractions, respectively. The migration of the molecular mass markers (expressed in kDa) is indicated. An arrow is used to demark PMP57 (see text). Migration of the peroxisomal proteins urate oxidase (UOX), catalase (CAT), and the inducible multifunctional protein (MFP) are also indicated (see Materials and Methods).
Ab-MF3 Inhibits PTS1-Protein Import.
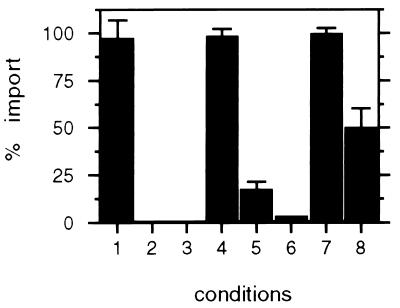
To analyze the role of PMP57 in peroxisome protein import, we measured the effect of ab-MF3 on HSA-PTS1 import into the peroxisomes of semiintact CHO cells (21, 27). To ensure the reproducible and characteristic behavior of the assay, we examined a number of conditions (see Materials and Methods for a description of our quantitation and normalization procedures). Several previously described properties of peroxisomal protein import were reproduced faithfully in this assay: (i) HSA-SKL import was unaffected by the presence of a 75-fold molar excess of the peptide CRYHLKPLQLKS (Fig. 2, condition 1), but was competed by a similar concentration of the PTS1-peptide CRYHLKPLQSKL (Fig. 2, condition 2), (ii) import of HSA-SKL into peroxisomes required an energy regenerating system (Fig. 2, condition 3), and (iii) both anti-hsp70 antibodies (Fig. 2, condition 6) and anti-HsPex5p antibodies (Fig. 2, condition 5) blocked HSA-SKL import into peroxisomes, in contrast to preimmune serum (Fig. 2, condition 4). With ab-MF3, but not with the corresponding preimmune serum, we consistently obtained approximately 50% HSA-PTS1 import inhibition (Fig. 2, conditions 8 versus 7, respectively). The specificity of this inhibition is indicated by the fact that under similar conditions antibodies to PMP70, an abundant peroxisomal membrane protein, fail to inhibit import (data not shown). This result implicates the involvement of PMP57 in peroxisomal protein import.
Figure 2.
Ab-MF3 inhibits PTS1-protein import in vitro. CHO cells were cultivated on coverslips, permeabilized with streptolysin-O, and incubated with HSA-SKL in transport buffer under various conditions, essentially as described (21). After washing and fixing, the coverslips were further processed by indirect immunofluorescence of HSA-SKL and quantitated as described in Materials and Methods. The results are expressed as the percentage of cells with peroxisomal HSA-SKL compared with the control standard. (1) + 75 molar excess CRYHLKPLQLKS (n = 2), (2) + 75 molar excess CRYHLKPLQSKL (n = 3), (3) − ATP-regenerating mix (n = 3), (4) + preimmune anti-HsPex5p (n = 2), (5) + anti-HsPex5p (n = 3), (6) + anti-hsp70 (n = 1), (7) + preimmune ab-MF3 (n = 3), and (8) + ab-MF3 (n = 3). (n refers to the number of separate experiments in which at least 250 cells in this condition were analyzed.)
Identification of PMPs That Bind HsPex5p in Ligand Blots.
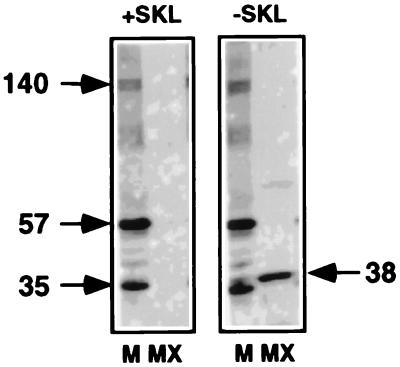
Ligand blotting was used to identify peroxisomal membrane proteins that have all the characteristics of bona fide docking proteins (Fig. 3). Rat liver peroxisomal membrane proteins as well as peroxisomal matrix proteins were prepared and separated by SDS/PAGE. The proteins were then transferred to nitrocellulose and renatured, and the membrane was incubated with a reticulocyte lysate containing [35S]HsPex5p. At the end of the incubation, the membranes were washed and subjected to autoradiography. Two proteins (57 and 35 kDa) were found to bind strongly to HsPex5p in an interaction that persisted in the presence of an excess of the CRYHLKPLQSKL peptide (Fig. 3). More weakly interacting species at other molecular weights, for example, at 140 kDa, also were seen occasionally, but these are not considered further here. The 57- and 35-kDa proteins are unlikely to be PTS1-containing proteins because (i) they are not recognized by antibodies directed against a mixture of matrix proteins (ab-MF16) or by anti-SKL antibodies (data not shown), (ii) the interaction of these proteins with Pex5p is not competed by the PTS1-peptide (Fig. 3), and (iii) under the same ligand-blotting conditions, only a 38-kDa peroxisomal matrix protein was recognized by Pex5p, but this binding was competed by the PTS1-peptide (Fig. 3).
Figure 3.
Pex5p-docking proteins in peroxisomal membranes. Ligand blotting of rat liver peroxisomal (M) membrane and (MX) matrix proteins in the presence (+SKL) or absence (−SKL) of 60 μM CRYHLKPLQSKL was performed as described in Materials and Methods. The migration of certain specific proteins is indicated with molecular masses, expressed in kDa.
The 57- and 35-kDa proteins appeared to be related, in a manner akin to the ab-MF3 immunoreactive species described above. Specifically, in some preparations, the 57-kDa protein was almost completely broken down to a 35-kDa species (and other lower-molecular-mass species) (data not shown). This similarity in molecular masses as well as the pattern of fragmentation suggest that we are looking at the same proteins.
Cloning of Human PEX14 and PEX13.
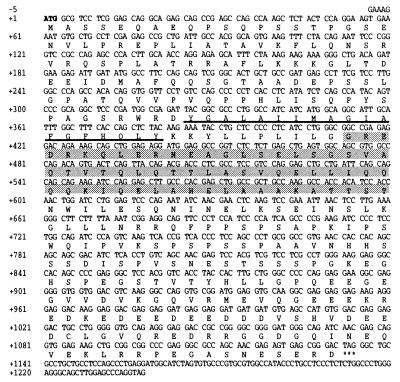
To determine whether the 57-kDa species that bound HsPex5p in the ligand blots is the human homologue of one of the recently described docking proteins, Pex14p or Pex13p, we expressed these proteins in a bacterial expression system. The cDNAs coding for HsPex14p and HsPex13p were obtained by probing the human EST database with HpPex14p/ScPex14p (9, 15) and HsPex13p (13), respectively. For HsPex14p, we found one human EST (GenBank accession no. H16035) encoding a protein homologous to both ScPex14p and HpPex14p. Sequence analysis revealed an ORF (GenBank accession no. AF045186) encoding a protein of 377 aa (Fig. 4) with a predicted molecular mass of 41 kDa. The deduced protein sequence displayed 21.7 and 24.1% identity to ScPex14p and HpPex14p, respectively (data not shown). Hydropathy analysis suggested that this protein contains, like ScPex14p and HpPex14p, one hydrophobic region (Fig. 4), but no membrane-spanning segments. In addition, we found a putative coiled-coil region (Fig. 4). Taken together, these results suggest that the protein described here can be considered as a good candidate for the human homologue of Pex14p. However, the human Pex14p did not contain the class II SH3-binding motif (28). In addition, the carboxyl terminus of HsPex14p is highly enriched in charged, largely acidic, amino acids (Fig. 4). Also, the expasy protparam tool program (Geneva, Switzerland) classified HsPex14p as an unstable protein (instability index: 73.25).
Figure 4.
Nucleotide sequence of the HsPEX14 cDNA and deduced amino acid sequence of HsPex14p. The putative start codon, indicated in bold, is surrounded by a perfect match to the Kozak consensus for mammalian initiator codons (29). Asterisks indicate the stop codon. The hydrophobic region is underlined. A putative coiled-coil region is shaded.
Probing the database with HsPex13 revealed, surprisingly, the existence of a human EST (GenBank accession no. AA295033) that encoded, in addition to the HsPex13p sequence reported (13), an additional 39 amino-terminal amino acids (GenBank accession no. AF048755). Taking into account that the initiation codon of this additional stretch is surrounded by a perfect match to the Kozak consensus for mammalian initiator codons (29), and that the proline-rich stretch of 39 aa is conserved in Pex13p of S. cerevisiae, this upstream methionine is the more likely candidate for initiation of translation.
HsPex14p Is Recognized Specifically by ab-MF3 and Binds HsPex5p and HsPex13p(SH3) in Ligand Blots.
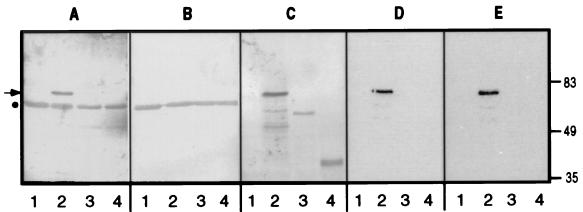
We probed the bacterial lysates containing recombinant (biotinylated-) HsPex14p with ab-MF3 and found a 70-kDa immunoreactive species (Fig. 5A, lane 2) (the aberrant migration of this molecule is considered below). This protein, which was not recognized by the ab-MF3 preimmune serum (Fig. 5B), was identified as (biotinylated-) HsPex14p by virtue of its recognition by streptavidin alkaline phosphatase (Fig. 5C, lane 2). No proteins were recognized specifically by ab-MF3 in lysates containing no expressed protein, the (biotinylated-) PTS1-binding domain of HsPex5p (30–32) or the (biotinylated-) SH3 domain of HsPex13p (Fig. 5A, lanes 1, 3, and 4). Importantly, the ab-MF3 immunoreactive species bound [35S]HsPex5p in ligand blots (Fig. 5D, lane 2) in a binding reaction not competed by the PTS1 peptide (Fig. 5E, lane 2). Also, no binding of biotinylated HsPex5p or of the biotinylated SH3 domain of HsPex13p to [35S]HsPex5p was observed (Fig. 5 D and E, lanes 3 and 4), although both of these molecules are present in the bacterial lysates (Fig. 5C, lanes 3 and 4).
Figure 5.
Recombinant HsPex14p is recognized specifically by ab-MF3 and binds HsPex5p in a ligand blot. Equal amounts of bacterial lysates expressing no recombinant protein (1), biotinylated HsPex14p (2), the biotinylated PTS1-binding domain of HsPex5p (3), or the biotinylated SH3 domain of HsPex13p (4) were subjected to SDS/PAGE and transferred to nitrocellulose. These membranes were then probed with ab-MF3 (A), preimmune serum (B), streptavidin-alkaline phosphatase (C), or [35S]Pex5p in the absence (D) or presence (E) of 60 μM CRYHLKPLQSKL, and reactive bands were visualized as described under Materials and Methods. The migration of molecular mass markers (in kDa) is indicated. The arrow indicates the recombinant expressed, biotinylated HsPex14p; the dot indicates the migration of a nonspecific, immunoreactive bacterial protein (A and B).
Considering the presence of a biotin tag that adds approximately 13 kDa to the molecular mass of proteins, we estimate HsPex14p to have an apparent molecular mass of 57 kDa. This observation was confirmed with a HsPex14p molecule to which a polyhistidine tag (approximately 1 kDa) was added and that was found to migrate indistinguishably from PMP57 (data not shown).
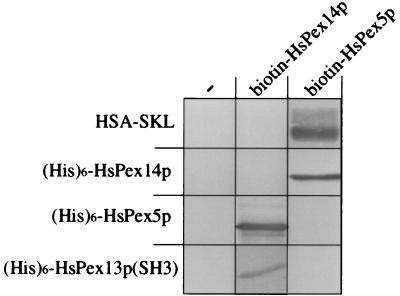
We further examined the binding properties of HsPex14p, HsPex13p(SH3), and HsPex5p through a series of pairwise incubations. Specifically, we immobilized different recombinant, purified proteins on nitrocellulose and examined their interaction with bacterially expressed biotinylated HsPex14p or HsPex5p (Fig. 6). We found, as expected, that biotinylated HsPex14p bound (His)6-HsPex5p and, conversely, biotinylated HsPex5p bound (His)6-HsPex14p. We also observed biotinylated HsPex5p binding to the PTS1 protein, HSA-SKL. Although we once again observed no interaction of (biotinylated) HsPex5p with (His)6-HsPex13p(SH3), we did, however, find biotinylated HsPex14p interacting, albeit weakly, with this protein. This important observation establishes an additional functional property of HsPex14p, that of interacting with HsPex13p.
Figure 6.
Interaction between HsPex5p, HsPex13p(SH3), and HsPex14p. Two micrograms of HSA-SKL or purified (His)6-tagged versions of HsPex14p, HsPex5p, and HsPex13p(SH3) was subjected to SDS/PAGE and transferred to nitrocellulose. Individual membranes containing these proteins then were incubated with a bacterial lysate containing no recombinant protein (−), biotinylated HsPex14p, or biotinylated HsPex5p (as indicated), and reactive complexes were identified with streptavidin-alkaline phosphatase. (For clarity, only the relevant portions of the nitrocellulose membranes are shown.)
DISCUSSION
The work described herein represents a description and functional characterization of Pex14p in mammalian cells. Using biochemical approaches, we identified human Pex14p as a peroxisomal membrane protein that binds the PTS1 receptor and is required for the import of PTS1-containing proteins into mammalian peroxisomes. This role is in concordance with studies in yeasts demonstrating that pex14Δ cells are deficient in the import of peroxisomal matrix proteins (9, 14, 15).
We have demonstrated earlier that the cytosol dependence of protein import into peroxisomes of permeabilized mammalian cells can be used to define cytosolic import factors, e.g., hsp70 (33). However, until now, the feasibility of using this in vitro import system for the identification of peroxisomal membrane components essential for import had not been determined. To address the role of peroxisomal membrane proteins, we raised antibodies against them and showed that one particular preparation, ab-MF3, inhibited HSA-SKL import (Fig. 2). This result clearly establishes the validity of an approach in which inhibiting antibodies are identified and used to define components of the import machinery.
Interestingly, although the inhibition we observe is specific, it is not as pronounced as that realized with antibodies to the largely cytosolic proteins, hsp70 or HsPex5p (Fig. 2). Several potential explanations may be considered, including that ab-MF3, although specific (Fig. 1), is of low titer. In addition, PMP57 may be only incompletely recognized in its native state in the context of the peroxisomal membrane. A less likely explanation is that there exists a PMP57-independent import process.
Biochemical fractionation of rat liver homogenates revealed the presence of an ab-MF3 immunoreactive 57-kDa protein in peroxisomes (Fig. 1). This protein is induced by the peroxisome proliferator clofibrate and behaves like a peroxisomal integral membrane protein or a polypeptide that is tightly associated with these membranes.
Ligand blotting of sodium carbonate-insoluble peroxisomal membrane proteins with radiolabeled HsPex5p showed clearly that proteins of 57 and 35 kDa bound the HsPex5p (Fig. 3). The molecular masses of these two polypeptides, and the observation that PMP57 in peroxisomal membrane fractions as well as the 57-kDa Pex5p-binding protein in the ligand blots were unstable species yielding a similar 35-kDa degradation product, implicated PMP57 as the Pex5p-binding protein. The interaction between PMP57 and HsPex5p is unlikely to be mediated by a PTS1 sequence on PMP57 because it is not competed by an excess of the canonical PTS1 peptide, under conditions where the interaction of a matrix protein (presumably urate oxidase) was (Fig. 3). It should be noted that the ligand blot conditions were clearly optimal for the binding of HsPex5p to membrane proteins because, unlike previously employed binding conditions (18), only one of the many PTS1-containing matrix proteins bound HsPex5p. Which specific components of the binding buffers facilitate the interaction with membrane proteins but not most matrix proteins remains an interesting, open question.
Two yeast Pex5p-binding proteins localized to the peroxisomal membrane were considered as potential candidates for homologues of mammalian PMP57. One of these, Pex13p, interacts with Pex5p via its cytosolically oriented, carboxyl-terminal SH3 domain (11–13). The second candidate is Pex14p, which, in addition to interactions with Pex5p, interacts with Pex7p, Pex13p, Pex17p, and itself (2, 9, 14, 15). A third potential candidate, Pex17p, was not considered because, although it is localized to the peroxisomal membrane and interacts with Pex5p in the yeast two-hybrid system, the latter binding is believed to be indirect and mediated by Pex14p (2).
To date, no mammalian homologues have been published for Pex14p, whereas a human homologue has been reported for Pex13p (13). We therefore cloned and sequenced the human orthologues of PEX14 and PEX13 (Fig. 4 and GenBank accession no. AF048755, respectively). Interestingly, the PEX13 cDNA was found to encode a protein 39 aa longer than reported previously (13).
Four independent lines of evidence prove that Pex14p and PMP57 are identical proteins. (i) Both the biotinylated, full-length HsPex14p expressed in bacteria and PMP57 are recognized by ab-MF3. (ii) Both polyhistidine-tagged Pex14p expressed in bacteria and rat liver PMP57 comigrate at 57 kDa (data not shown). (iii) Both Pex14p and PMP57 bind radiolabeled HsPex5p in ligand blots. (iv) The interaction between these proteins and HsPex5p is unchanged by the presence of the PTS1 peptide. Because ab-MF3 recognizes both of these proteins and inhibits peroxisomal matrix protein import, these results document the essential functional role of Pex14p in import by serving as a Pex5p docking site on the peroxisomal membrane.
In view of published reports that yeast Pex5p and the SH3 domain of Pex13p interact, our inability to detect binding between HsPex5p and the SH3 domain of HsPex13p may seem surprising (Figs. 5 and 6). Why we are unable to detect this interaction is unclear. The HsPex13p(SH3) molecule renatures and is active, at least partially, because we can detect an interaction with HsPex14p.
With respect to published reports regarding the function of yeast Pex14p and Pex5p, our demonstration of the peroxisomal membrane location of mammalian Pex14p, its interaction with Pex5p and Pex13p, and its involvement in the import of peroxisomal matrix proteins reinforces the theme that components involved in peroxisomal protein import and biogenesis are conserved through evolution. Further characterization of this connection is critical for a molecular understanding of the basis of the human peroxisomal disorders. We are investigating whether or not any human peroxisomal disorders are caused by mutations in HsPex14p.
Acknowledgments
M.F. is a postdoctoral fellow of the Fonds voor Wetenschappelijk Onderzoek–Vlaanderen. This work also was funded by National Institutes of Health Grant DK41737 to S.S.
ABBREVIATIONS
- PTS
peroxisomal targeting signal
- SH3
Src homology 3
- EST
expressed sequence tag
- PMP
peroxisomal membrane protein
Footnotes
References
- 1.Distel B, Erdmann R, Gould S J, Blobel G, Crane D I, Cregg J M, Dodt G, Fujiki Y, Goodman J M, Just W W, et al. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau W-H. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau W-H, Erdmann R. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramani S. Physiol Rev. 1998;78:171–187. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- 5.Rachubinski R A, Subramani S. Cell. 1995;83:525–528. doi: 10.1016/0092-8674(95)90091-8. [DOI] [PubMed] [Google Scholar]
- 6.Gould S J, Keller G A, Subramani S. J Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swinkels B, Gould S J, Bodnar A G, Rachubinski R A, Subramani S. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- 9.Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel J A K W, Veenhuis M, Kunau W-H. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- 10.Lazarow P B, Fujiki Y. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 11.Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak H F, Distel B. J Cell Biol. 1996;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdmann R, Blobel G. J Cell Biol. 1996;135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould S J, Kalish J E, Morrell J C, Bjorkman J, Urquhart A J, Crane D I. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brocard C, Lametschwandtner G, Koudelka R, Hartig A. EMBO J. 1997;16:5491–5500. doi: 10.1093/emboj/16.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komori M, Rasmussen S W, Kiel J A K W, Baerends R J S, Cregg J M, van der Klei I J, Veenhuis M. EMBO J. 1997;16:44–53. doi: 10.1093/emboj/16.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verheyden K, Fransen M, Van Veldhoven P P, Mannaerts G P. Biochim Biophys Acta. 1992;1109:48–54. doi: 10.1016/0005-2736(92)90185-o. [DOI] [PubMed] [Google Scholar]
- 17.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fransen M, Brees C, Van Veldhoven P P, Mannaerts G P. Anal Biochem. 1996;242:26–30. doi: 10.1006/abio.1996.0423. [DOI] [PubMed] [Google Scholar]
- 19.Alconada A, Gartner F, Honlinger A, Kubrich M, Pfanner N. Methods Enzymol. 1995;260:263–286. doi: 10.1016/0076-6879(95)60144-9. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 21.Wendland M, Subramani S. J Cell Biol. 1993;120:675–685. doi: 10.1083/jcb.120.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuervo A M, Dice J F. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 23.Wiemer E A C, Nuttley W M, Bertolaet B L, Li X, Francke U, Wheelock M J, Anne U K, Johnson K R, Subramani S. J Cell Biol. 1995;130:51–65. doi: 10.1083/jcb.130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rapp S, Soto U, Just W W. Exp Cell Res. 1993;205:59–65. doi: 10.1006/excr.1993.1058. [DOI] [PubMed] [Google Scholar]
- 28.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 29.Kozak M. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brocard C, Kragler F, Simon M M, Schuster T, Hartig A. Biochem Biophys Res Commun. 1994;204:1016–1022. doi: 10.1006/bbrc.1994.2564. [DOI] [PubMed] [Google Scholar]
- 31.Fransen M, Brees C, Baumgart E, Vanhooren J C T, Baes M, Mannaerts G P, Van Veldhoven P P. J Biol Chem. 1995;270:7731–7736. doi: 10.1074/jbc.270.13.7731. [DOI] [PubMed] [Google Scholar]
- 32.Terlecky S R, Nuttley W M, McCollum D, Sock E, Subramani S. EMBO J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walton P A, Wendland M, Subramani S, Rachubinski R A, Welch W J. J Cell Biol. 1994;125:1037–1046. doi: 10.1083/jcb.125.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]