Abstract
The ability to specifically target a mitogenic signal to a population of genetically modified primary cells would have potential applications both for gene and cell therapy. Toward this end, a gene encoding a fusion protein containing the FK506-binding protein FKBP12, fused to the intracellular portion of the receptor for thrombopoietin (mpl), was introduced into primary murine bone marrow cells. Dimerization of this fusion protein through the addition of a dimeric form of the drug FK506, called FK1012, resulted in a marked proliferative expansion of marrow cells that was restricted to the genetically modified population. FK1012’s proliferative effect was sustained and reversible. An apparent preference for differentiation along the megakaryocytic lineage was observed. This approach allows for the specific delivery of a mitogenic signal to a population of genetically modified primary cells and may have applications for studies in hematopoiesis and receptor biology, and for gene and cell therapy.
Stem cell gene therapy is constrained by the inefficiency of gene transfer into early hemopoietic progenitors and stem cells. One strategy for surmounting this obstacle is to expand the population of genetically modified cells through selection. When this approach is used, a gene encoding a selectable product is coupled to a gene encoding a therapeutic protein. After selection, cells containing the therapeutic gene emerge (1). Selection may be applied ex vivo (1) or, if a clinically tolerable regimen were devised, repeated cycles of selection might be undertaken in vivo (2). Conventional methods for selection involve the transfer of a drug-resistance gene followed by exposure to the corresponding cytotoxic drug. The applicability of this approach is limited by two factors. First, cytotoxic drugs have undesired consequences when administered in vivo. Second, the persistent enrichment of genetically modified cells is expected to require that selection be exerted at the level of early hematopoietic cells. However, selective pressure is very difficult to apply at the level of progenitors and stem cells because of their intrinsic resistance to killing by most cytotoxic agents (3, 4).
An alternative method for achieving selection would be to provide the genetically modified cell population with a direct growth advantage. The practical application of this strategy would require that the growth advantage: (i) be restricted to the genetically modified cell population and (ii) be completely reversible. We have reported recently the development of a method that appears to meet these requirements (5). This method uses a system that permits intracellular protein dimerization to be reversibly activated in response to a lipid-soluble dimeric form of the drug FK506, called FK1012 (6–13). FK1012 is used as a pharmacological mediator of dimerization to bring together two FK506-binding domains taken from the endogenous protein FKBP12. Fusion proteins composed of FKBP12 linked to the signaling domain of the erythropoietin receptor allowed Ba/F3 cells, which are interleukin 3 (IL-3)-dependent, to be switched to FK1012 dependency (5). The studies described herein were performed to determine whether FK1012 can be used for the selection and ex vivo expansion of genetically modified primary murine bone marrow cells.
MATERIALS AND METHODS
Retroviral Producer Lines.
Retrovirus producer lines were generated by transfection of the amphotropic packaging line PA317 (14) with vector plasmid, followed by supernatant collection 2 days later that was subsequently used for transduction of the ecotropic packaging line GpE+86 (15). Producer clones were isolated in the presence of G418 and screened for virus titer based on end-point titration using NIH 3T3 cells (16). Producer clones were isolated for each vector and yielded titers between 105 and 106 colony-forming units/ml. Genetic stability was confirmed by Southern analysis of clones and pools of producer cells by using the restriction enzyme KpnI, which cuts once in each long terminal repeat, and a probe directed against the neo gene. Producer lines were documented to be free of helper-competent virus by using a marker-rescue assay (17). Cell lines were cultured at 37° in DMEM (GIBCO/BRL, Life Technologies, Grand Island, NY) supplemented with 8% fetal calf serum (FCS; GIBCO/BRL), sodium pyruvate, nonessential amino acids (GIBCO/BRL), and 2 mM l-glutamine (GIBCO/BRL).
Retroviral Transduction.
Fluorouracil, 150 mg/kg, was injected intraperitoneally into female B6D2F1 mice. Forty-eight hours later, marrow cells were harvested and cultured for 48 hr in DMEM containing 16% FCS, 5% IL-3-conditioned medium (Collaborative Biomedical Products, Bedford, MA), 100 ng/ml recombinant human IL-6, and 50 ng/ml recombinant murine stem cell factor (Peprotech, Rocky Hill, NJ) in a 37°C, 5% CO2 incubator. After 48 hr of prestimulation, cells were transferred onto irradiated (1,500 cGy) producer cells and cocultivated by using identical growth conditions except for the addition of polybrene (8 μg/ml). Marrow cells were harvested after 48 hr of cocultivation.
Suspension Cultures.
After retroviral transduction, marrow cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) containing 10% fetal calf serum (FCS), 50 units/ml penicillin, 50 μg/ml streptomycin either in the presence or absence of FK1012 (100 nM) or flt-3 ligand (100 ng/ml) (Peprotech), in a 37o, 5% CO2 incubator. Cell numbers were determined on the days indicated.
Clonogenic Assays.
Cells were plated at a concentration of 5 × 103 cells/ml in cultures containing 30% FCS, 1% BSA, 5 × 10−4 M 2-mercaptoethanol (Sigma), 10% WEHI-3 cell-conditioned medium, and 1% methylcellulose (Fisher Scientific). Transduction efficiency was assessed by using G418 (GIBCO/BRL) at an active concentration determined to be sufficient to kill all nontransduced cells (800 μg/ml). Cultures were plated in triplicate and incubated in a highly humidified 37°C 5% CO2 incubator. Colonies were evaluated on day 8.
RESULTS
FK1012-Mediated Dimerization of the mpl Cytoplasmic Domain Stimulates Proliferative Signaling.
In an attempt to generate a signaling molecule that is capable of stimulating growth among early hemopoietic cells, constructs containing the signaling domain of mpl were tested (Fig. 1). Mpl, the receptor for thrombopoietin (18–20), was selected because of thrombopoietin’s demonstrated capacity for stimulating proliferation in multilineage hemopoietic progenitors in vivo (21) and because of the small size of the mpl signaling domain, which facilitates incorporation into retroviral vectors. Two constructs were tested (Fig. 1A). F3mpl encodes a membrane-targeted fusion protein containing three copies of the FKBP12 motif fused to the intracellular portion of mpl, whereas the protein encoded by F1mpl differs in that it contains only a single FKBP12 motif. Ba/F3 cell clones stably expressing either of these constructs were tested, after IL-3 withdrawal, for the capacity to proliferate in response to FK1012 (7) (Fig. 1B). Two clones expressing the F3mpl construct and a single clone expressing the F1mpl construct all exhibited dose-dependent proliferative responses in the presence of FK1012. An alternative dimerizer of FKBP12 domains called AP1510 (22, 23) stimulated proliferation in both clones expressing F3mpl but failed to significantly induce proliferation in the clone expressing F1mpl (data not shown). Clones expressing either the F3mpl or F1mpl constructs remained dependent on the presence of either dimerizer or IL-3 for survival (data not shown).
Figure 1.
Test of constructs. (A) Schematic design. An XhoI linkered fragment encoding the intracellular domain of murine c-mpl was PCR-amplified by using Pfu polymerase and the following primer pairs: 5′-GGCTCGAGAAGTGGCAATTTCCTGCG-3′ and 5′-GGCTCGAGGGGCTGCTGCCAATAGC-3′. After sequence confirmation, the XhoI-digested fragment was inserted into the SalI site of the construct F3 (5) to produce F3mpl. F3mpl contains a myristylation domain to direct localization to the inner surface of the cell membrane, three copies of the FK506-binding peptide FKBP12, the intracellular portion of c-mpl, and a hemagglutinin epitope tag to permit detection of the fusion protein by Western blot assay. F1mpl differs from F3mpl in that it contains only a single FKBP12 site. These constructs were used to generate Ba/F3 clones expressing high levels of the fusion protein as described previously (5). (B) Cell proliferation (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, MTT) assays for transfected Ba/F3 cell clones expressing either F3mpl (solid symbols) or F1mpl (open circles) were performed as described previously (5). Peak proliferative responses were observed at a concentration of 100 nM FK1012. (C) MTT assays for retrovirally transduced Ba/F3 cell clones expressing F1mpl driven by MSCVneo EB (24). Results, plotted as a fraction of the OD570–630 obtained in the same clones using 5% IL-3-containing WEHI conditioned medium, denote the mean of three experiments. Error bars denote standard deviations.
FK1012 Stimulates Expansion of Genetically Modified Primary Bone Marrow Cells.
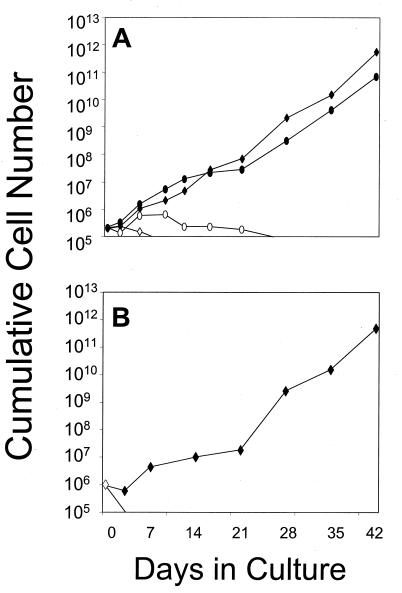
To test whether FK1012-mediated activation of mpl is capable of inducing proliferation in genetically modified bone marrow cells, the F1mpl construct was inserted into the retroviral vector MSCV-neoEB (24). The resulting construct, MSCVF1mpl, was used to generate a stable, high-titer, helper virus-free ecotropic producer cell line. Supernatant from the MSCVF1mpl producer cell line was capable of conferring G418 resistance and FK1012 responsiveness in transduced Ba/F3 cell clones (Fig. 1C). Gene transfer into murine bone marrow cells was performed by cocultivation (16), and transduced marrow cells were tested in suspension culture for the ability to respond to FK1012 (Fig. 2). FK1012 was evaluated both in the absence of added growth factors (Fig. 2), and in the presence of flt-3 ligand (FL) (Fig. 2A), because thrombopoietin and FL exert synergistic effects on hemopoietic cell expansion ex vivo (25). Cells were counted at various times during culture. In the absence of drug or added growth factors, all nonadherent cells died within 7 days, whereas adherent macrophages persisted for a variable period of time (Fig. 2 and data not shown). The presence of FL alone produced a 3-fold expansion of cells (Fig. 2A); however, all cells died by day 28 of culture. In contrast, FK1012 dramatically stimulated cell proliferation (Fig. 2). The proliferative effect of FK1012 was apparent within 5 days of culture and by 42 days in culture had resulted in a 2.5 million-fold expansion of cells in one experiment (Fig. 2A), and a near 500,000-fold cell expansion in a second experiment (Fig. 2B). The combination of FK1012 and FL resulted in nearly equivalent rates of cell growth compared with FK1012 alone (Fig. 2A).
Figure 2.
FK1012 stimulates expansion of genetically modified bone marrow cells. Results from two independent experiments (A and B) are shown. After retroviral transduction, marrow cells were cultured in IMDM containing 10% FCS, in the presence or absence of FK1012 (100 nM) (A and B) or flt-3 ligand (100 ng/ml) (A). Cells were counted at various times during culture. (⋄) No FK1012, no FL; (♦) FK1012 alone; (○) FL alone; (•) FK1012 plus FL. Note that the y axis is on a logarithmic scale.
Temporal Evolution of FK1012-Responsive Cell Lineages.
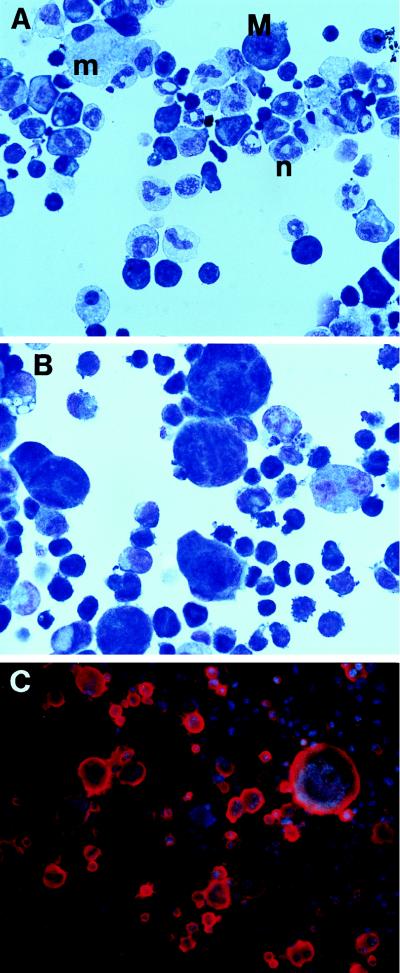
The types of cells emerging in response to FK1012 differed at varying time points of culture (Fig. 3). During the first 2 weeks of culture in FK1012, mature granulocytes and macrophages predominated, whereas megakaryocytes were less frequent (Fig. 3A). At later time points mature granulocytes fell dramatically and megakaryocytes emerged as the dominant phenotypically identifiable cell population (Fig. 3 B and C). Similar changes in culture over time were also noted in the experiment depicted in Fig. 2B (data not shown). Morphologically similar findings also were observed with the combination of FK1012 plus FL, with the exception that macrophages were more abundant than with FK1012 alone (data not shown).
Figure 3.
FK1012-responsive cell lineages. (A) Wright–Giemsa stain after 9 days of culture in FK1012. Neutrophils (n) and macrophages (m) are abundant. Small numbers of megakaryocytes (M) also can be seen. (B) Wright–Giemsa stain after 42 days of culture in FK1012. Neutrophils have disappeared, and megakaryocytes emerge as the predominant phenotypically identifiable cell population. (C) Anti-CD41 alkaline phosphatase staining of cells cultured for 42 days in FK1012. Megakaryocyte cytoplasm stains red and nuclei stain blue. Note that the full spectrum of megakaryocyte maturation is observed, from small, blast-like cells to large, polyploid cells.
To demonstrate that the proliferative effect of FK1012 is reversible, cells cultured for 28 days in the presence of FK1012 were tested for persistence of cell growth after FK1012 withdrawal. As shown in Table 1, withdrawal of FK1012 was followed promptly by cell death, whereas readdition of FK1012 was associated with persistent cell growth.
Table 1.
Reversibility of FK1012-dependent cell proliferation
| Growth conditions | Day in culture
|
||
|---|---|---|---|
| 0 | 7 | 14 | |
| +FK1012 | 5 | 36 | 1,283 |
| −FK1012 | 5 | 0.59 | .09 |
After 28 days of culture in FK1012 (100 nM), transduced marrow cells were washed extensively and cultured in suspension in the presence (+) or absence (−) of FK1012. Numbers indicate cells per well × 10−5.
FK1012 Stimulates Expansion Among Clonogenic Progenitor Cells.
To determine whether FK1012 is capable of stimulating expansion among clonogenic progenitor cells, progenitors were assayed at various time points of culture as colony-forming unit cells (CFU-C) (3, 4). As the MSCVF1mpl vector incorporates a neo gene (24), the frequency of genetically modified progenitors is reflected in the frequency of G418-resistant CFU-C. In the absence of FK1012, CFU-C expansion failed to occur, and there was no preferential survival advantage in favor of G418-resistant CFU-C. By day 28 CFU-C had fallen to undetectable levels (Fig. 4A). In contrast, in the presence of FK1012 an almost 600,000-fold expansion in CFU-C was observed by day 42 (Fig. 4B). The vast majority of CFU-C were G418-resistant, suggesting a preferential proliferative advantage in favor of the genetically modified progenitor cell population. Of note is that although granulocytes were virtually absent after 42 days in suspension culture in the presence of FK1012 alone (Fig. 2B), CFU-C progeny derived from these cultures contained large numbers of granulocytes (data not shown). Because CFU-C assays were performed in the presence of IL-3, these findings suggest that myeloid progenitors arising in response to FK1012 require additional growth factors to accomplish terminal differentiation. Similar effects of FK1012 were observed in the presence of FL. FL alone produced a transient 7-fold expansion in CFU-C without preference for the transduced population (Fig. 4C and data not shown). In contrast, the combination of FL and FK1012 produced a 260,000-fold expansion of transduced CFU-C by day 42 (Fig. 4D). Very similar levels of progenitor cell expansion were observed in the second experiment depicted in Fig. 2B (data not shown).
Figure 4.
CFU-C assays at various time points during suspension culture. Cells were harvested on the days indicated and cultured in semisolid media either in the presence or absence of G418. The concentration of G418, 800 μg/ml, was sufficient to prevent the growth of nontransduced cells (data not shown). Solid bars, CFU-C numbers in the presence of G418; open bars, CFU-C numbers in the absence of G418. (A) No FK1012, no FL. (B) FK1012 alone. (C) FL alone. (D) FK1012 plus FL. Note that FK1012-mediated CFU-C expansion markedly favors the genetically modified cell population.
Although the data presented above suggest that FK1012 selectively expands genetically modified progenitor cells, an assessment of preferential expansion is complicated by high baseline transduction rates into progenitors (consistently >80%). To determine whether FK1012 is capable of specifically delivering a mitogenic signal to a minor population of genetically modified CFU-C, a third experiment was performed in which marrow cells cocultivated with the MSCVF1mpl producer cell line were mixed with marrow cells cocultivated with the parental packaging cell line. The frequency of G418-resistant CFU-C arising from this mixed population of transduced and nontransduced cells was 35%. As shown in Table 2, FK1012 selectively expanded the genetically modified progenitor cell population.
Table 2.
FK1012-mediated selection of genetically modified CFU-C
| Day in culture | No drug | FK1012 | FL | FK1012 + FL |
|---|---|---|---|---|
| 0 | 0.35 | 0.35 | 0.35 | 0.35 |
| 7 | 0.24 | 0.96 | 0.14 | 0.98 |
Numbers indicate fraction of G418-resistant CFU-C.
DISCUSSION
Data presented herein demonstrate that FK1012-mediated dimerization of a fusion protein containing the intracellular signaling domain of mpl is capable of stimulating proliferation in genetically modified primary hematopoietic cells. Results indicate that the mitogenic signal delivered by FK1012 is specifically targeted to the genetically modified cell population. The ex vivo application of this approach may be relevant to the expansion of genetically modified cell populations including megakaryocytes, other cell lineages, and possibly stem cells. The in vivo application of this approach may allow for the specific expansion of a minor population of genetically modified stem cells and progenitors and thus may be useful for gene therapy.
In addition to the potential utility of this system for therapeutic applications, the results obtained may provide insights into blood cell development. During the first 2 weeks of culture, FK1012 stimulated the expansion of multiple cell lineages, including granulocytes and macrophages. In contrast, at later time points of culture FK1012 directed the expansion of a cell population that was dominated by megakaryocytes with very few mature granulocytes. Despite the dramatic decline in mature granulocyte production, progenitors with the potential for granulocytic differentiation were retained, as evidenced by the emergence of granulocytes upon culture in the presence of IL-3. FK1012’s ability to stimulate terminal granulocytic maturation during the first 2 weeks of culture contrasts sharply with the loss of mature granulocytes at later time points. Among possible explanations for this phenomenon is that granulocytes arising in response to FK1012 at early time points in culture were derived from progenitors preprogrammed to a myeloid pathway of differentiation, and that signaling through mpl permitted their survival. In contrast, progenitors generated under the influence of FK1012 may require signals other than those provided by mpl to carry out granulocytic differentiation. The apparent inability of mpl to support sustained granulocytic maturation is supported by a recent study using the full-length mpl receptor (26).
Equally noteworthy is the predominance of megakaryocytes at later time points in the culture. The absence of thrombopoietin in these cultures was demonstrated by the failure of supernatant to support the growth or survival of Ba/F3 cells expressing the full-length mpl receptor (data not shown). Megakaryocytic differentiation also has been observed frequently in cell lines generated using v-mpl (27). These findings suggest that mpl activation preferentially enables differentiation along the megakaryocytic lineage. We presently are unable to distinguish whether this phenomenon arises from the preferential survival and expansion of megakaryocyte progenitors, or whether mpl signaling influences lineage specification in multipotential progenitor cells (28). Using this system, it may be possible to delineate mpl subdomains that are necessary for megakaryocytic differentiation.
In aggregate, these findings demonstrate that FK1012-mediated activation of the mpl signaling domain is capable of stimulating expansion among genetically modified megakaryocytes, hematopoietic progenitor cells, and possibly stem cells. This system may provide important insights into stem cell and receptor biology and may have applications for gene and cell therapy.
Acknowledgments
We thank Janis Abkowitz and Virginia Broudy for helpful discussions, Denise Farrar for technical assistance, and Michael Gilman (Ariad Pharmaceuticals) for providing FK1012 and AP1510. This work was supported by Grants 1R01 DK52997–01, 5P01 HL53750, and 5P30 DK47754 from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: IL, interleukin; CFU-C, colony-forming unit cells.
References
- 1.Migita M, Medin J A, Pawliuk R, Jacobson S, Nagle J W, Anderson S, Amiri M, Humphries R K, Karlsson S. Proc Natl Acad Sci USA. 1995;92:12075–12079. doi: 10.1073/pnas.92.26.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorrentino B P, Brandt S J, Bodine D, Gottesman M, Pastan I, Cline A, Nienhuis A W. Science. 1992;257:99–103. doi: 10.1126/science.1352414. [DOI] [PubMed] [Google Scholar]
- 3.Blau C A, Neff T, Papayannopoulou T. Hum Gene Ther. 1996;7:2069–2078. doi: 10.1089/hum.1996.7.17-2069. [DOI] [PubMed] [Google Scholar]
- 4.Blau C A, Neff T, Papayannopoulou T. Blood. 1997;89:146–154. [PubMed] [Google Scholar]
- 5.Blau C A, Peterson K R, Drachman J G, Spencer D M. Proc Natl Acad Sci USA. 1997;94:3076–3081. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 7.Pruschy M N, Spencer D M, Kapoor T M, Miyake H, Crabtree G R, Schreiber S L. Chem Biol. 1994;1:163–172. doi: 10.1016/1074-5521(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 8.Spencer D M, Graef I, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9805–9809. doi: 10.1073/pnas.92.21.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holsinger L J, Spencer D M, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9810–9814. doi: 10.1073/pnas.92.21.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer D M. Trends Genet. 1996;12:181–187. doi: 10.1016/0168-9525(96)10013-5. [DOI] [PubMed] [Google Scholar]
- 12.Ho S N, Biggar S, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 13.Spencer D M. Curr Biol. 1996;6:839–847. doi: 10.1016/s0960-9822(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 14.Miller A D, Buttimore C. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markowitz D, Goff S, Bank A. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodine D M, McDonagh K T, Brandt S J, Ney P A, Agricola B, Byrne E, Nienhuis A W. Proc Natl Acad Sci USA. 1990;87:3738–3742. doi: 10.1073/pnas.87.10.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller A D, Rosman G J. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 18.Lok S, Kaushansky K, Holly R D, Kuijper J L, Lofton-Day C D, Oort P J, Grant F J, Hepel M D, Burkhead S K, Kramer J M, et al. Nature (London) 1994;369:565–568. doi: 10.1038/369565a0. [DOI] [PubMed] [Google Scholar]
- 19.Kaushansky K, Lok S, Holly R D, Broudy V C, Lin N, Bailey M C, Forstrom J W, Buddle M M, Oort P J, Hage F S, Roth G J, Papayannopoulou T, Foster D C. Nature (London) 1994;369:568–571. doi: 10.1038/369568a0. [DOI] [PubMed] [Google Scholar]
- 20.Wendling F, Maraskovsky E, Debili N, Florindo C, Teepe M, Titeux M, Methia N, Breton-Gorius J, Cosman D, Vainchenker W. Nature (London) 1994;369:571–574. doi: 10.1038/369571a0. [DOI] [PubMed] [Google Scholar]
- 21.Kaushansky K, Lin N, Grossmann A, Humes J, Sprugel K H, Broudy V C. Exp Hematol. 1996;24:265–269. [PubMed] [Google Scholar]
- 22.Amara J F, Clackson T, Rivera V M, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage N L, Holt D A, Gilman M. Proc Natl Acad Sci USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin L, Asano H, Blau C A. Blood. 1998;91:890–897. [PubMed] [Google Scholar]
- 24.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 25.Piacibello W, Sanavio F, Garetto L, Severino A, Bergandi D, Ferrario J, Fagioli F, Berger M, Aglietta M. Blood. 1997;89:2644–2653. [PubMed] [Google Scholar]
- 26.Goncalves F, Lacout C, Villeval J L, Wendling F, Vainchenker W, Dumenil D. Blood. 1997;89:3544–3553. [PubMed] [Google Scholar]
- 27.Souyri M, Vigon I, Pencioelli J-F, Heard J-M, Tambourin P, Wendling G. Cell. 1990;63:1137–1147. doi: 10.1016/0092-8674(90)90410-g. [DOI] [PubMed] [Google Scholar]
- 28.Cross M A, Enver T. Curr Opin Genet Dev. 1997;7:609–613. doi: 10.1016/s0959-437x(97)80007-x. [DOI] [PubMed] [Google Scholar]