Studies of the involvement of ECM molecules in cell attachment, growth, and differentiation have revealed a central role of heparan sulfate (HS) proteoglycans (HSPGs) in early embryogenesis, morphogenesis, angiogenesis, and epithelial-mesenchymal interactions (1–3). HS chains bind a multitude of proteins and ensure that a wide variety of bioactive molecules (e.g., heparin-binding growth factors, chemokines, lipoproteins, and enzymes) cling to the cell surface and ECM. HSPGs can thus influence a variety of normal and pathologic processes, among which are tissue repair, neurite outgrowth, inflammation and autoimmunity, tumor growth and metastasis, vasculogenesis and angiogenesis (1–4). Binding to HS can modulate a tethered molecule’s biological activity or protect it from proteolytic cleavage and inactivation. Transmembrane and phospholipid-anchored HSPGs (syndecans and glypicans, respectively) mediate cell interactions with components of the microenvironment that control cell shape, adhesion, proliferation, survival, and differentiation (2, 3). These species of HSPGs can also serve as coreceptors along with the other cell surface molecules to form functional receptor complexes that transduce signals from various ligands (2, 3).
Because of the important and multifaceted roles of HSPGs in cell physiology, their cleavage is likely to alter the integrity and functional state of tissues and to provide a mechanism by which cells can respond rapidly to changes in the extracellular environment. Enzymatic degradation of HS is, therefore, likely to be involved in fundamental biological phenomena, ranging from pregnancy, morphogenesis, and development to inflammation, angiogenesis, and cancer metastasis. Contrary to some early claims, there is no good evidence for more than one endogenous mammalian HS-degrading endoglycosidases. The recent cloning of a single gene by several groups (5–10), together with biochemical studies (11), suggests that mammalian cells express primarily a single dominant heparanase enzyme (12). This Perspective focuses on the molecular properties, expression pattern, biological functions, and clinical significance of this heparanase in normal and pathological processes, with emphasis on tumor metastasis and angiogenesis.
Molecular and biochemical properties of heparanase
HS degradation by mammalian endoglycosidic enzymes was first described in human placenta and rat liver hepatocytes. Since then, heparanase activity has been identified in a variety of normal and malignant cells and tissues, among which are cytotrophoblasts, endothelial cells (ECs), platelets, mast cells, neutrophils, macrophages, T and B lymphocytes, and lymphoma, melanoma, and carcinoma cells (5, 6, 12–16). Heparanase cleaves the glycosidic bond with a hydrolase mechanism and is thus distinct from bacterial heparinases, which depolymerize heparin and HS by eliminative cleavage. HS glycosaminoglycan chains are cleaved by heparanase at only a few sites, yielding HS fragments of appreciable size (10–20 sugar units), suggesting that the enzyme recognizes a particular and relatively rare HS structure (17).
Heparanase purified from human platelets and hepatoma requires the presence of O-sulfation, with no essential requirement for N-sulfation or IdoA residues (17). The heparin-derived octasaccharide, which binds antithrombin III is cleaved at a single site (Figure 1, top, arrow), indicating that a 2-O-sulfate group on a hexuronic acid residue located two monosaccharides away from the cleavage site is essential for substrate recognition by heparanase (17) (Figure 1). In other studies, however, the presence of either N- or O-sulfates did not appear to be an absolute requirement for substrate cleavage (12, 18).
Figure 1.
Heparanase-mediated release of bioactive molecules sequestered in the ECM. Heparanase cleaves HSPGs (arrows) and releases a variety of physiologically and pathologically important molecules. Inset shows the cleavage site within the antithrombin-binding heparin 3H-octasaccharide. The actual antithrombin-binding sequence corresponds to sugar units 2–6 (within brackets). X in sugar unit 4 represents hydrogen or SO–3. Adapted from Pikas et al. (17).
The cloning of a single human heparanase cDNA sequence and its expression in mammalian cells was independently reported by several groups (5–10). The heparanase cDNA contains an open reading frame of 1629 bp encoding a 61.2-kDa polypeptide of 543 amino acids. The mature active 50 kDa enzyme, isolated from cells and tissues, has its N-terminus 157 amino acids downstream from the initiation codon (5–12), suggesting post-translational processing of the heparanase polypeptide at an unusual cleavage site (Gln157-Lys158). Processing and activation occur during incubation of the full-length 65-kDa recombinant enzyme with several normal and transformed cells and, to a lesser extent, with their conditioned medium (5). The putative cell surface proteinase that activates the latent heparanase has not been characterized, but preliminary studies indicate that the heparanase precursor may bind to the cell surface, most likely to HS, and is then converted to its highly active 50 kDa form in a process accompanied by endocytosis of the processed form (Katz, B.-Z., et al., our unpublished results). Heparanase activity is readily obtained after transfection of mammalian cells with cDNAs encoding the entire heparanase precursor (5–10). Attempts to express the truncated 50-kDa (Lys158 to Ile543) protein failed, however, to yield active enzyme, suggesting that regions N-terminal to Lys158 are required for expression and/or function of the protein. In fact, the active enzyme has been postulated to be a heterodimer of the 50-kDa subunit noncovalently associated with an 8-kDa peptide (Gln36 to Glu109), which arises from proteolytic processing of the pre-proheparanase protein (9). It has not been determined whether association of the 50-kDa polypeptide with the 8-kDa fragment is essential for heparanase activity (9).
The predicted amino acid sequence of heparanase includes six putative N-glycosylation sites, five of which cluster in the first 80 amino acids of the 50-kDa mature protein. Removal of N-glycosylation does not affect the enzyme activity (5). The sequence also contains a putative N-terminal signal peptide sequence (Met1 to Ala35) and a candidate transmembrane region (Pro515 to Ile534) (5, 6, 12). Alignment of the human, mouse, and rat heparanase amino acid sequences corresponding to the 50-kDa human mature enzyme (Lys158 to Ile543) revealed 80–93% identity (6); 61% homology was found between the recently cloned chicken heparanase (19) and the human enzyme. A prominent difference between the chicken and the mammalian enzymes resides in their signal peptide sequence, accounting for the chicken enzyme being secreted and localized in close proximity to the cell surface. In contrast, the human heparanase is mostly intracellular, localized in perinuclear granules (19).
The fact that highly homologous cDNA sequences were derived from different species and types of normal and malignant cells is consistent with the notion that one dominant HS-degrading endoglycosidase is expressed by mammalian cells (5–10, 12). Thus, unlike the large number of proteases that can solubilize polypeptides in the ECM, one major heparanase appears to be used by cells to degrade the HS side chains of HSPGs. A putative heparanase 2, which shows 35% homology with the heparanase 1 described above, was recently cloned, although no enzymatic activity has been associated with this gene product (20). Unlike heparanase 1, heparanase 2 mRNA expression shows a wide distribution in normal tissues (20).
Secondary structure predictions suggest that heparanase contains an (α/β)8 TIM-barrel fold (residues 411–543), characteristic of the clan A glycosyl hydrolase families (11). Site-directed mutagenesis reveals that, as with other TIM-barrel glycosyl hydrolases, heparanase’s catalytic mechanism involves two conserved acidic residues, a putative proton donor at Glu225 and a nucleophile at Glu343. Conserved basic residues are found in proximity to the proposed catalytic proton donor (Lys231 and Lys232) and nucleophile (Lys337 and Lys338) (11). The heparanase gene (∼50 kb) is located on human chromosome 4q21.3 and is linked to the genetic marker D4S400 (21). The gene is expressed as 5 kb (HPSE 1a) and 1.7 kb (HPSE 1b) mRNA species, generated by alternative splicing. The HPSE 1a form contains 14 exons and 13 introns, whereas in HPSE 1b, the first and fourteenth exons have been spliced out (21).
Regulation of heparanase activity
Because of the potential tissue damage that could result from inadvertent cleavage of HS, heparanase must be tightly regulated, although little is known about the control of its expression, activity, or subcellular localization. Heparanase activity has been detected in both chloroquine-sensitive (lysosomal) and -insensitive (endosomal) compartments of rat ovarian and human colon carcinomas. The enzyme has been localized in perinuclear acidic endosomal and lysosomal granules of fibroblasts and tumor cells (Katz, B.-Z., et al., our unpublished results) and in the tertiary granules of human neutrophils, where it is colocalized with matrix metalloproteinase-9 (MMP-9) (16, 22). It will be of interest to determine whether these molecules are coordinately regulated in various cellular contexts.
Several observations suggest that heparanase can be membrane-bound. Heparanase immunoreactivity is observed on the surface of various human cancer cells, including colon adenocarcinoma (23). The heparanase sequence contains a putative hydrophobic transmembrane domain (11, 12), and its complete solubilization from rat liver, platelets, and tumor cells, requires the presence of a detergent, indicating that up to 25% of the heparanase activity in these preparations is associated with the membrane (6). Interestingly, mannose-6-phosphate displaces heparanase from the surface of T lymphocytes, suggesting that it binds to surface-expressed mannose-6-phosphate receptor (6, 12). Bioinformatic analysis of the protein sequence also predicts that heparanase can exist as a glycosylphosphatidylinositol-linked protein on the surface of cells.
Soluble heparanase exhibits maximal endoglycosidase activity between pH 5.0 and 6.0 and is inactivated at pH greater than 8.0. Cell surface bound heparanase is moderately active at pH 6.7, but the nonvascularized core of tumor masses might provide the acidic environment required for heparanase degradation of ECM. At physiological pH, where very little enzymatic activity is evident, heparanase binds to HSPGs, where it may facilitate leukocyte adhesion and extravasation in response to inflammatory conditions (24).
Preferential expression of heparanase in human tumors
Expression of the human heparanase mRNA in normal tissues is restricted primarily to the placenta and lymphoid organs (5, 6, 20). Immunohistochemistry shows that heparanase occurs primarily in neutrophils, macrophages, platelets, cytotrophoblasts, keratinocytes, capillary endothelium, and neurons, with little or no staining in connective tissue cells and most normal epithelia.
As judged by quantitative RT-PCR, heparanase mRNA is increased in human malignancies and xenografts of human breast, colon, lung, prostate, ovary, and pancreas tumors, compared with the corresponding normal tissues (20). Heparanase mRNA and protein accumulate even at early stages in the progression of human colon carcinoma, and their levels increase gradually as cells progress from severe dysplasia through well-differentiated to poorly differentiated colon carcinoma; adjacent, morphologically normal colonic tissue shows no expression of the enzyme. Deeply invading colon carcinoma cells and the adjacent desmoplastic stromal fibroblasts show high levels of the heparanase mRNA and protein (23).
Human mammary carcinomas likewise express the heparanase mRNA and protein in both the in situ and the invasive components of ductal and lobular origins. Breast carcinoma cells that have entered the circulation and lymph node metastases show particularly intense immunostaining, whereas normal breast tissue expresses little or no heparanase (5, 25). Preferential expression of the heparanase mRNA and protein in tumors is also evident in tissue specimens derived from adenocarcinoma of the ovary, metastatic melanoma, oral squamous cell carcinoma, hepatocellular carcinoma, and carcinomas of prostate, bladder, and pancreas (refs. 5, 23, 26, and our unpublished observations). In addition, enhanced heparanase mRNA expression correlates significantly with reduced postoperative survival of patients with pancreatic cancer (26).
Heparanase in tumor metastasis and angiogenesis
HSPGs are prominent components of blood vessels. In large vessels they are concentrated mostly in the intima and inner media, whereas in capillaries they are found mainly in the subendothelial basement membrane (BM), where they support proliferating and migrating ECs and stabilize the structure of the capillary wall. Cleavage of HS is therefore expected to facilitate extravasation of blood-borne tumor cells, as well as sprouting of angiogenic ECs (Figure 2). It was originally thought that the role of ECM-degrading enzymes is to break down tissue barriers, thus enabling tumor cells to invade through stroma and blood vessel walls. In recent years it has been appreciated that extravasation of blood-borne tumor cells may not be the primary rate-limiting step in the metastatic cascade and that matrix degradation enzymes — MMPs and heparanase among them — not only promote cell invasion but also induce angiogenesis by modulating growth factor activity and bioavailability.
Figure 2.
Heparanase-mediated extravasation of blood-borne cells. Heparanase expressed by tumor cells (left) and neutrophils (right) promotes cell invasion between adjacent vascular ECs and through their underlying basal lamina (BL) into the ECM. Platelets may facilitate extravasation of blood-borne cells through binding to the invading cells, and activation and release of their own heparanase enzyme. Left: Scanning electron micrographs showing invasion of T-lymphoma cells, in the absence (top) or presence (bottom) of platelets, through a monolayer of cultured vascular ECs.
Cancer invasion and metastasis involves degradation of ECM constituents, including collagens, laminins, fibronectin, vitronectin, and HSPGs. The malignant cell is able to accomplish this task through the concerted sequential action of enzymes such as MMPs, serine and cysteine proteases, and endoglycosidases. Expression of heparanase correlates with the metastatic potential of human tumor cells (5, 6, 12–14). Moreover, elevated levels of heparanase have been detected in sera of animals and human cancer patients bearing metastatic tumors and in the urine of some patients with aggressive metastatic disease (ref. 13, and our unpublished observations). In addition, Andela et al. recently found that inhibition of NF-κB signaling coordinately downregulates MMP-9, plasminogen activator, and heparanase and thus prevents experimental and spontaneous metastasis (27). Conversely, coculture with astrocytes, or treatment of metastatic brain melanoma cells with nerve growth factor or neurotrophin-3, stimulates both heparanase activity and cell invasion, suggesting that astrocytes may significantly contribute to melanoma brain colonization (28).
Remarkably, heparanase-inhibiting molecules, such as nonanticoagulant species of heparin, polysulfated polysaccharides, and other polyanionic molecules, reduce the incidence of experimental metastases by more than 90% (12–15, 18). Evidence for a direct role of heparanase in tumor metastasis is seen in the conversion of T-lymphoma cells from nonmetastatic to metastatic behavior following stable transfection and overexpression of heparanase (5). A massive liver infiltration of the transfected cells and accelerated mortality of the mice were observed following subcutaneous inoculation of heparanase-overexpressing cells, compared with mice inoculated with mock-transfected lymphoma cells. Similarly, transient transfection with a heparanase cDNA increases lung colonization of intravenously inoculated mouse melanoma cells (5). These effects were best demonstrated with a secreted, membrane-bound form of heparanase (ref. 19; Goldshmidt, O., our unpublished results).
Angiogenesis represents a coordinated multicellular process involving a wide variety of molecules, including growth factors, ECM components, adhesion receptors, and matrix-degrading enzymes. HSPGs and HSPG-degrading enzymes have long been implicated in cell invasion, migration, adhesion, differentiation, and proliferation (2, 3, 29), all processes that are associated with angiogenesis. Heparin and HS sequester, stabilize, and protect FGFs and VEGFs from inactivation. Moreover, these molecules can function as low-affinity coreceptors that promote the formation of HS-FGF-FGFR complexes, thus facilitating receptor dimerization and signaling (see Iozzo and Antonio, this Perspective series, ref. 30; and refs. 2, 3, 29, 31) (Figure 3). Both recombinant and native heparanase expressed by platelets, tumor, and inflammatory cells release an active complex of bFGF and an HS fragment from ECM and BM (4, 31). The length of HS required for stimulation of FGF-receptor binding and dimerization is similar to that of HS fragments released by heparanase (17, 31).
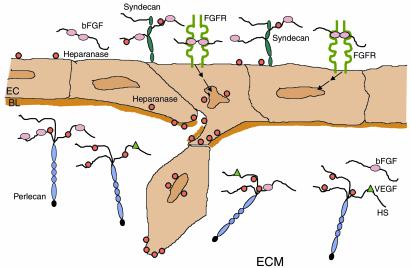
Figure 3.
Proposed involvement of heparanase in angiogenesis. Heparanase promotes: EC migration and degradation of the subendothelial basal lamina (BL) and ECM; release of active HS-bound bFGF and VEGF; and release of HS degradation fragments that promote FGF-receptor (FGFR) binding, dimerization, and signaling (arrows), inducing EC migration and proliferation.
An important early step in the angiogenic cascade is degradation of the subendothelial capillary BM by proliferating ECs, a prerequisite for the formation of vascular sprouts. By degrading the polysaccharide scaffold of BM, heparanase may facilitate EC invasion and migration toward angiogenic factors (Figure 3), much as MMPs and other proteolytic enzymes are presumed to do. We have demonstrated a high expression of heparanase mRNA in proliferating human ECs. Moreover, immunohistochemical staining of human colon and breast carcinomas revealed preferential expression of the heparanase protein by ECs of sprouting capillaries in the vicinity of the tumor, with little or no staining of mature vessels (32). Using the mouse Matrigel plug angiogenesis assay, we observed an increased angiogenic response to heparanase-transfected T-lymphoma cells, embedded in Matrigel and implanted subcutaneously; mock-transfected control cells elicited no such response (32). Similarly, MMP-9 is now regarded as a specific component of the angiogenic switch, because it renders VEGF more available to its receptors and its inhibitors impair angiogenesis and tumor growth (33).
Heparanase in normal development and tissue remodeling
Most studies emphasize the involvement of heparanase in pathophysiology. Little is known about the enzyme contribution to normal cell and tissue function. However, we have observed that heparanase mRNA and protein are specifically expressed in the developing chick, in cells migrating from the epiblast and forming the hypoblast layer, as early as 6 hours after fertilization. Later, by 72 hours, the enzyme is preferentially expressed in cells of the developing vascular and nervous systems (19). Heparanase might prove to be essential for embryo implantation through its effects on cellular invasive properties (10). The roles heparanase plays in the adult are likely to include wound repair, tissue regeneration, and immune surveillance. Clearly, the development of mice with targeted disruption of the heparanase gene is needed to elucidate its normal roles in embryonic development and in the mature individual.
An intact BM is essential for the proper function, differentiation, and morphology of many epithelia. Disruption or loss of BM occurs during normal development and in the disease state. To investigate the involvement of heparanase in tissue remodeling and differentiation, we have generated transgenic mice that overexpress the heparanase mRNA and protein in all tissues. The most pronounced phenotype was noted in the mammary gland. Mammary glands taken from heparanase-overexpressing virgin females show precocious alveolar development and maturation, with primary and secondary ducts, similar to those of a normal pregnant mouse (25). Intact BM is inhibitory for growth and sprouting of epithelial cells both in vivo and in vitro. Heparanase-dependent cleavage of BM HSPGs appears to disrupt this physical barrier and to unmask ECM molecules and liberate HS-bound growth and differentiation factors, such as bFGF. Extracellular proteolysis is required for branching morphogenesis (34), as revealed by the highly differentiated morphological and functional phenotype of mammary glands of transgenic mice overexpressing stromelysin-1 (34), which also show a high incidence of premalignant or malignant phenotypes, ranging from severe hyperplasia to adenocarcinoma (34). Our preliminary results suggest that overexpression of heparanase may be associated with similar alterations.
Inhibitory molecules and clinical considerations
Nonanticoagulant species of heparin and various sulfated polysaccharides — fucoidan, pentosan sulfate, carrageenan-λ, laminaran sulfate — that inhibit experimental metastasis also inhibit tumor cell heparanase. Intriguingly, other polymers (e.g., chondroitin sulfate, carrageenan-κ, hyaluronic acid) have little or no effect on both parameters (12–15). While the mechanism underlying this correlation is in need of study, treatment with low molecular weight heparin has been observed to confer a lower mortality rate on cancer patients (35).
We (14) have observed that heparin species containing more than ten sugar units and having sulfate groups at both the N and the O positions are the most potent both at inhibiting heparanase activity and at blocking experimental metastasis. While O-desulfation abolished the inhibitory effect of heparin, replacement of N-sulfates by N-acetyl or N-hexanoyl groups had only a small effect on the inhibitory activity (14). Potent inhibition of heparanase activity and tumor metastasis has also been demonstrated with other heparin-mimicking compounds and polyanionic molecules, although an effect on selectin-mediated cell adhesion cannot be excluded as the mechanism for these beneficial effects (35).
Parish et al. (18) have initiated a comprehensive screening program to identify sulfated oligosaccharides that can inhibit tumor metastasis and promote tumor regression by their effects on heparanase activity and angiogenic growth factor action. Oligosaccharide chain length and degree of sulfation emerged as more important parameters than the sugar composition and type of linkage in this study. With increasing sulfation there was a steady increase in the ability of maltohexaose to inhibit both heparanase activity and experimental metastasis (18). Phosphomannopentaose sulfate (PI-88) and maltohexaose sulfate were comparable to heparin in their inhibitory activity (IC50 1–2 μg/ml). Continuous administration of PI-88 inhibits growth, vascularity and lymph node metastasis of mammary adenocarcinoma tumors in rats. This compound is being evaluated in a multicenter phase II clinical trial (12, 18).
Competitive inhibition of heparanase by PI-88 and other sulfated oligosaccharides and modified heparin derivatives may also be applied to suppress autoimmune and chronic inflammatory diseases. However, the pleiotropic effects and interactions of such heparin/HS mimetics with heparin-binding proteins might elicit undesirable effects. Random, high-throughput screening of chemical libraries, preparation of neutralizing antibodies, and rational design of compounds that block the heparanase active site or ligand-binding domain are among the other approaches applied to develop effective heparanase inhibitors. Natural endogenous heparanase inhibitors may also be identified.
Concluding remarks
Tumor spread involves degradation of macromolecules in the ECM and blood vessel wall. Among these are HSPGs, which play a key role in the self-assembly, solubility, and barrier properties of BM and ECM, as well as in sequestration and stabilization of bioactive molecules. Expression of heparanase correlates with the metastatic potential of tumor cells, and treatment with heparanase inhibitors markedly reduces the incidence of metastasis in experimental animals. bFGF and VEGF, which bind heparin and promote angiogenesis, are stored in the microenvironment of tumors, most often as a complex with HS. These proteins are released and can induce new capillary growth when HS is degraded by heparanase. In fact, overexpression of heparanase elicits an angiogenic response in vivo (32). Thus, apart from its involvement in tumor spread, heparanase may be a component of the angiogenic switch, promoting tumor vascularization and growth. Hence, ECM-degrading enzymes (e.g., MMPs and heparanase) affect cell migration and spread, as well as tumor growth at primary and secondary sites.
Given the potential tissue damage that could result from inadvertent cleavage of HS, tight regulation and balance are essential. Potential regulators are cytokines, hormones, local pH, and cellular localization. Regulatory elements in the promoter and other regions of the heparanase gene are being investigated, although very little is known at the present time. An attractive regulatory target is the putative membrane-bound proteinase that converts heparanase from a latent into an active form. The nature of this enzyme is not known. The significance of cell surface expression and secretion of the enzyme, its activation, cellular uptake, and the associated effects on cell migration, metastasis, and angiogenesis are being investigated.
It appears that activated T lymphocytes use the same enzymatic machinery as do tumor cells to traverse the vascular endothelium, reach their target tissue, and elicit inflammation and autoimmune disorders (16). Whereas heparanase expression is induced upon activation of cells of the immune system, the enzyme is constitutively expressed by acute myeloid leukemia, but not chronic lymphocytic leukemia (our unpublished results). Its role in platelets is unknown. Investigation of the expression of heparanase in hematopoietic cells may better elucidate the control of its expression and its potential role in normal differentiation, mobilization, and homing of bone marrow–derived cells.
Clearly, heparanase offers an attractive drug target. The unexpected identification of a single predominant functional heparanase suggests that if its activity can be inhibited, there may be no other enzymes available to compensate for its loss. On the other hand, taking into account the normal functions of the enzyme, heparanase-inhibiting compounds might interfere with normal functions such as immune surveillance, tissue repair, anticoagulant activity, and HS turnover. Species of heparin and heparin/HS-mimicking compounds that inhibit the enzyme may, for example, displace HS-bound growth-promoting factors and thereby elicit undesirable angiogenesis. These compounds may shift the balance from free bFGF to HS-bound bFGF and hence enhance cellular responses to bFGF and other heparin-binding growth factors.
It is hoped that identification of the sugar residues in HS adjacent to the heparanase cleavage site, as well as crystallization and analysis of the three dimensional structure of the enzyme, will lead to a rational design of highly specific heparanase inhibitors. Several groups are currently developing competitive heparin/HS-mimicking compounds, neutralizing anti-heparanase antibodies, antisense oligonucleotides, and heparanase-inhibiting small molecules. Conversely, because heparanase itself promotes cell migration and angiogenesis, administration and/or upregulation of the enzyme in vivo may prove useful to accelerate wound healing and neovascularization.
Heparanase is the first mammalian HS-degrading enzyme that has been cloned, expressed, and characterized. This work may pave the way for identification and cloning of other mammalian glycosaminoglycan-degrading enzymes (e.g., chondroitinase, dermatanase, and keratanase) and thus represent a step toward a better understanding of the function and biological significance of these enzymes and their polysaccharide substrates.
Acknowledgments
We gratefully acknowledge the help and support of Iris Pecker, Dror Melamed, and other colleagues at InSight Ltd. (Rabin Science Park, Rehovot, Israel). The devotion, motivation, and continuous assistance of our Tumor Biology Research team are highly appreciated. This work was supported by grants from the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities; the Israel Cancer Research Fund; the Association for International Cancer Research, United Kingdom; the NIH (R21 CA87085); and the US Army (grant 0278).
References
- 1.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Bernfield M, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 4.Vlodavsky, I., Bar-Shavit, R., Korner, G., and Fuks, Z. 1993. Extracellular matrix-bound growth factors, enzymes and plasma proteins. In Basement membranes: cellular and molecular aspects. D.H. Rohrbach and R. Timpl, editors. Academic Press. Orlando, Florida, USA. 327–343.
- 5.Vlodavsky I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 6.Hulett MD, et al. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 7.Kussie PH, et al. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261:183–187. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 8.Toyoshima MT, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153–24160. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- 9.Fairbanks MB, et al. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem. 1999;274:29587–29590. doi: 10.1074/jbc.274.42.29587. [DOI] [PubMed] [Google Scholar]
- 10.Dempsey LA, Brunn GJ, Platt JL. Heparanase, a potent regulator of cell matrix interactions. Trends Biochem Sci. 2000;25:349–355. doi: 10.1016/s0968-0004(00)01619-4. [DOI] [PubMed] [Google Scholar]
- 11.Hulett MD, et al. Identification of active-site of the pro-metastatic endoglycisidase heparanase. Biochemistry. 2000;39:15659–15667. doi: 10.1021/bi002080p. [DOI] [PubMed] [Google Scholar]
- 12.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima M, Irimura T, Nicolson GL. Heparanases and tumor metastasis. J Cell Biochem. 1988;36:157–167. doi: 10.1002/jcb.240360207. [DOI] [PubMed] [Google Scholar]
- 14.Vlodavsky I, et al. Inhibition of tumor metastasis by heparanase inhibiting species of heparin. Invasion Metastasis. 1994;14:290–302. [PubMed] [Google Scholar]
- 15.Parish CR, Coombe DR, Jakobsen KB, Bennett FA, Underwood PA. Evidence that sulphated polysaccharides inhibit tumour metastasis by blocking tumour-cell-derived heparanases. Int J Cancer. 1987;40:511–518. doi: 10.1002/ijc.2910400414. [DOI] [PubMed] [Google Scholar]
- 16.Vlodavsky I, et al. Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis. 1992;12:112–127. [PubMed] [Google Scholar]
- 17.Pikas DS, Li JP, Vlodavsky I, Lindahl U. Substrate specificity of heparanases from human hepatoma and platelets. J Biol Chem. 1998;273:18770–18777. doi: 10.1074/jbc.273.30.18770. [DOI] [PubMed] [Google Scholar]
- 18.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 19.Goldshmidt O, et al. Expression pattern and secretion of human and chicken heparanase are determined by their signal peptide sequence. J Biol Chem. 2001;276:29178–29187. doi: 10.1074/jbc.M102462200. [DOI] [PubMed] [Google Scholar]
- 20.McKenzie E, et al. Cloning and expression profiling of hpa2, a novel mammalian heparanase family member. Biochem Biophys Res Commun. 2000;276:1170–1177. doi: 10.1006/bbrc.2000.3586. [DOI] [PubMed] [Google Scholar]
- 21.Dong J, Kukula AK, Toyoshima M, Nakajima M. Genomic organization and chromosome localization of the newly identified human heparanase gene. Gene. 2000;253:171–178. doi: 10.1016/s0378-1119(00)00251-1. [DOI] [PubMed] [Google Scholar]
- 22.Mollinedo F, et al. Major co-localization of the extracellular-matrix degradative enzymes heparanase and gelatinase in tertiary granules of human neutrophils. Biochem J. 1997;327:917–923. doi: 10.1042/bj3270917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedmann Y, et al. Expression of heparanase in normal, dysplastic and neoplastic human colon mucosa and stroma. Am J Pathol. 2000;157:1167–1175. doi: 10.1016/S0002-9440(10)64632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilat D, et al. Molecular behavior adapts to context: heparanase functions as an extracellular matrix-degrading enzyme or as a T cell adhesion molecule, depending on the local pH. J Exp Med. 1995;181:1929–1934. doi: 10.1084/jem.181.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zcharia E, et al. Molecular properties and involvement of heparanase in cancer progression and mammary gland morphogenesis. Mammary Gland Biology & Neoplasm. 2001;6:311–322. doi: 10.1023/a:1011375624902. [DOI] [PubMed] [Google Scholar]
- 26.Koliopanos A, et al. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001;61:4655–4659. [PubMed] [Google Scholar]
- 27.Andela VB, Schwarz EM, Puzas JE, O’Keefe RJ, Rosier RN. Tumor metastasis and the reciprocal regulation of prometastatic and antimetastatic factors by nuclear factor κB. Cancer Res. 2000;60:6557–6562. [PubMed] [Google Scholar]
- 28.Marchetti D, Li J, Shen R. Astrocytes contribute to the brain-metastatic specificity of melanoma cells by producing heparanase. Cancer Res. 2000;60:4767–4770. [PubMed] [Google Scholar]
- 29.Folkman J, Shing Y. Control of angiogenesis by heparin and other sulfated polysaccharides. Adv Exp Med Biol. 1992;313:355–364. doi: 10.1007/978-1-4899-2444-5_34. [DOI] [PubMed] [Google Scholar]
- 30.Iozzo RV, San Antonio JD. Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J Clin Invest. 2001;108:349–355. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlodavsky I, Miao H-Q, Medalion B, Danagher P, Ron D. Involvement of heparan sulfate and related molecules in sequestration and growth promoting activity of fibroblast growth factor. Cancer Metastasis Rev. 1996;15:177–186. doi: 10.1007/BF00437470. [DOI] [PubMed] [Google Scholar]
- 32.Elkin M, et al. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 33.Bergers G, et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2:737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sternlicht MD, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borsig L, et al. Heparin and cancer revisited: mechanistic connections involving platelets, P-selectin, carcinoma mucins, and tumor metastasis. Proc Natl Acad Sci USA. 2001;98:3352–3357. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]