Abstract
The Notch receptor, which is involved in numerous cell fate decisions in invertebrates and vertebrates, is synthesized as a 300-kDa precursor molecule (p300). We show here that proteolytic processing of p300 is an essential step in the formation of the biologically active receptor because only the cleaved fragments are present at the cell surface. Our results confirm and extend recent reports indicating that the Notch receptor exists at the plasma membrane as a heterodimeric molecule, but disagree as to the nature of the protease that is responsible for the cleavage that takes place in the extracellular region. We report here that constitutive processing of murine Notch1 involves a furin-like convertase. We show that the calcium ionophore A23187 and the α1-antitrypsin variant, α 1-PDX, a known inhibitor of furin-like convertases, inhibit p300 processing. When expressed in the furin-deficient Lovo cell line, p300 is not processed. In vitro digestion of a recombinant Notch-derived substrate with purified furin allowed mapping of the processing site to the carboxyl side of the sequence RQRR (amino acids 1651–1654). Mutation of these four amino acids (and of two secondary dibasic furin sites located nearby) completely abolished processing of the Notch1 receptor.
The Notch gene family encodes transmembrane receptors of 300 kDa that are involved in cell fate choices in vertebrates and invertebrates (1, 2). The extracellular domain contains 10–36 copies of an epidermal growth factor (EGF)-like sequence motif and 3 copies of a Lin-12/Notch/Glp (LNG) sequence motif. The intracellular domains contain six to seven Ankyrin repeats and a PEST-containing region. Activation of Notch receptors upon ligand binding results in inhibition of differentiation and can also transduce inductive signals (1, 2). Ligands (Delta and Serrate), modulators of ligand activity (Fringe family), intracellular components [Deltex, Su(H)], and target genes have been identified in Drosophila and in vertebrates (2) but the biochemical events associated with Notch signaling so far have remained elusive. One model hypothesizes that ligand binding induces processing of Notch, followed by nuclear translocation of an intracytoplasmic fragment of the receptor, that associates with the Su(H)/RBP DNA-binding molecule to activate target genes (3–5). To investigate the biochemical and physiological relevance of this model we first studied the structure and subcellular localization of the mammalian Notch1 receptor in the absence of ligand. Western blot analyses of mammalian and Drosophila tissues have shown that, in addition to the 300-kDa form, a protein of approximately 120 kDa lacking most of the extracellular part is revealed (6, 7).
Indeed, Blaumueller et al. (7) recently have shown that the human Notch2 receptor is cleaved in the trans-Golgi network and suggest that the truncated protein of 110–120 kDa, which includes the intracellular domain, is associated with the extracellular domain to form a heterodimeric molecule able to bind the ligand Delta. Through use of a coimmunoprecipitation assay we formally demonstrate that, after cleavage, the extracellular part of the murine Notch1 receptor is associated with the intracellular part bound to the cell membrane. Pan and Rubin (8) proposed that the disintegrin metalloprotease encoded by the gene Kuzbanian (KUZ), which is required during lateral inhibition mediated by Notch in Drosophila neurogenesis (9), is involved in constitutive Notch receptor cleavage. They showed that expression of a dominant negative form of KUZ lacking the prodomain and the protease domain inhibited constitutive Notch processing and led to an increased number of neurogenic cells in Drosophila and Xenopus laevis, confirming that KUZ is necessary for the lateral inhibitory signal. Using different inhibitors of protease activity, and a cell line devoid of furin activity, we show that the constitutive processing of the murine Notch1 receptor is a result of a furin-like protease. This enzyme belongs to a family of subtilisin-like, calcium-dependent convertases that process proproteins in the constitutive secretory pathway (10). Finally by analysis of Notch1 mutants and in vitro digestion experiments we identified the cleavage site and confirmed that the Notch1 receptor is indeed constitutively cleaved by furin.
MATERIALS AND METHODS
Cells, Transfections, and Extract Preparations.
HeLa, LoVo, and 293T cells were grown in DMEM supplemented with 10% fetal calf serum. Jurkat cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. HeLa cells were stably transfected with the murine Notch1 cDNA cloned into the pcDNA3 vector (Invitrogen) after G418 selection (0.8 mg/ml). A clone was selected for further studies, and named HeLa(N1). Jurkat cells were stably transfected with the α1-PDX cDNA cloned into pcDNA3. One of the transformants was selected for further studies and named C12D. Transient and stable transfections were performed by calcium phosphate coprecipitation for 293T and HeLa cells, using lipofectamine (GIBCO/BRL) for LoVo cells and by electroporation [260V, 1500 μF, using a Cellject electroporator (Eurogentec, Brussels)] for Jurkat cells. For extract preparation, cells were washed with cold PBS and lysed in TNT lysis buffer [50 mM Tris⋅HCl, pH 8/200 mM NaCl/0.5% Nonidet-40/1 mM Pefabloc SC (Boehringer Mannheim)]. After 20 min on ice, cell lysates were centrifuged at 14,000 rpm for 20 min, and the supernatants were used for immunoprecipitation or Western blot analysis.
Antibodies.
IC antiserum. A DNA fragment encoding amino acids 1759–2306 of the intracellular domain of murine Notch1 was cloned by PCR into the vector pGEX-KT. The GST-Notch1 fusion protein was purified on a glutathione agarose column and injected into rabbits. The crude serum was purified on an Affigel column coupled to the fusion protein.
EC antiserum.
A peptide encoding amino acids 1621–1637 was coupled to KLH and injected into rabbits.
Immunoprecipitation of Metabolically Labeled Extracts.
Two days after transfection, HeLa and LoVo cells grown in 60-mm dishes (≈4 × 106 cells) were incubated in methionine-free DMEM for 2 hr. They were then labeled for 15 min with 200 μCi/ml of [35S]methionine and chased in complete medium supplemented with 2.5 mM methionine for different times (see Fig. 3). About 6 × 106 Jurkat T cells were treated as described for HeLa and LoVo cells. Cells were washed and extracts were prepared as described above. The supernatant was precleared with protein A-Sepharose (Sigma) and incubated with the purified IC antibody and protein A-Sepharose in RIPA buffer. The immunoprecipitates were washed and boiled in reducing gel sample buffer for subsequent analysis on SDS gels. When a second immunoprecipitation was performed, the immunocomplex was dissociated by boiling for 5 min in 50 mM Tris⋅HCl, pH 8/2% SDS diluted 20-fold in TNT buffer and incubated in the presence of the second antiserum and protein A-Sepharose.
Figure 3.
Notch1 processing does not take place in LoVo cells. HeLa cells (lanes 1–4) or LoVo cells, either transfected with Notch1 cDNA (lanes 5–8) or cotransfected with Notch1 and furin cDNAs (lanes 9–12) were labeled for 15 min with [35S]methionine and then chased for the indicated times. Extracts were prepared and immunoprecipitated with antiserum IC and analyzed on 6.5% SDS-polyacrylamide gels. Molecular mass markers (kDa) are indicated on the left of lane 5, and the positions of p300 and p120 are shown.
Cell Surface Labeling.
Jurkat cells (30 × 106) were labeled as described above with [35S]methionine (50 μCi/ml) for 2 hr and subsequently washed three times with PBS buffer. Sulfo-NHS-LS biotin (Pierce) was added to the cells at 1 mg/ml for 1 hr at 4°C. After incubation with PBS containing 0.15% glycine, the cells were washed and lysed in TNT buffer. The extracts were directly immunoprecipitated with IC antiserum and protein A-Sepharose in RIPA buffer or incubated with streptavidine-agarose beads in RIPA buffer for 2 hr at 4°C. The beads then were washed, and bound proteins were eluted with 50 mM Tris⋅HCl, pH 8/2% SDS for 5 min at 100°C, diluted 20-fold with TNT buffer, and immunoprecipitated in the presence of antiserum IC.
Western Blotting.
Cell extracts were subjected to SDS/PAGE and transferred (3 hr) onto Immobilon membranes (Millipore), which were incubated with the primary antibody (IC diluted 3,000-fold or EC 1,000-fold), followed by peroxidase-conjugated anti-rabbit Igs, and revealed by using the enhanced chemiluminescence system (Pierce).
Mutagenesis.
The target for mutagenesis was a 1,585-bp StuI-BglII mouse Notch1 fragment subcloned into pRc/CMV (Invitrogen). Site-directed mutagenesis was carried out by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Each mutant was sequenced to confirm the mutation, and mutated fragments were recloned in the Notch1 cDNA into pcDNA3. The oligonucleotide used to mutate RQRR to AAAA was GGTACCAGTGGTGGGGCGGCCGCTGTAGAGCTGGACCCCATG; for mutating the 2 dibasic sites, the oligonucleotide was CACGAGGAAGAGCTGGCTGCCCACCCAATCGCTGGCTCTACAGTGGGTTGGGCC. For this mutagenesis the target was a StuI-BglII mNotch1 fragment mutated first at the RQRR site.
In Vitro Digestion of Recombinant Notch1 Fusion Proteins by Furin and Microsequencing.
The mNotch1 fragments spanning amino acid 1564 to amino acid 1723, wild type or mutated on the RQRR site, were cloned by PCR into pGEX-KT. The 3′ oligonucleotide encoded 6 additional histidines. The fusion proteins were purified on glutathionine-agarose columns and dialyzed against 50 mM Tris⋅HCl, pH 7.4/1 mM CaCl2/0.5% Triton X-100. The digestion was performed in the same buffer in the presence of 100 units purified recombinant BCRD-furin. After overnight digestion at room temperature, the products were electrophoresed on a 15% SDS/Tricine gel, followed by silver staining. The C-terminal digestion product of the wild-type molecule was purified on nickel-agarose beads according to standard procedures. The purified protein (5 pmol) was blotted onto a poly(vinylidene difluoride) membrane. Sequencing was performed on an Applied Biosystems 473A sequencer.
RESULTS
Structure of the Notch1 Receptor at the Cell Surface.
The cDNA of the Notch1 receptor encodes a 300-kDa protein, but previous immunoblot analyses have revealed in many cells and tissues the presence of both the 300-kDa (hereafter referred to as p300) and a 120-kDa (p120) proteins, the former corresponding to the full-length molecule, and the latter including the intracellular region of the molecule, the transmembrane domain, and a small part of the extracellular region. Aster et al. (6) have shown that in Jurkat cells, a cell line derived from a T lymphoblastic tumor that expresses Notch1, the p120 protein, is generated by proteolytic cleavage of the p300 precursor. To examine whether p300 and/or p120 are expressed at the cell surface, Jurkat cells were labeled with [35S]methionine, mock-treated, or treated with biotin succinimidyl-ester, lysed and immunoprecipitated with an antiserum raised against the intracellular region of Notch1 (antiserum IC) either directly (Fig. 1A, lanes 1 and 3) or after adsorption on streptavidin-agarose (lanes 2 and 4). The only protein binding to streptavidin-agarose beads and immunoprecipitated by an antibody raised against the intracellular region of Notch1 is p120 (lane 4). This result suggests that the p120 cleavage product, but not the full-length precursor, can be transported to the cell surface. Identical results have been obtained for the Notch2 receptor (7). Because EGF repeats 10–11 are required for binding of the ligand to the receptor (11), to constitute a functional entity the p120 must be associated with the N-terminal remaining part of the receptor. A 200-kDa protein that apparently corresponds to the remaining extracellular domain of Notch has been detected (7, 12), but no direct evidence of association between the two processing products of p300, p120, and p200 has been provided so far. To detect such an association, we performed immunoprecipitation with the IC antiserum and analyzed the immunoprecipitate by Western blotting using an antibody raised against a peptide located in the extracellular domain of the receptor (amino acids 1621–1637: serum EC). Fig. 1B shows the results of immunoprecipitation of an extract derived from HeLa cells stably transfected with murine Notch1 cDNA [referred to as HeLa(N1)]. When serum IC is used for immunoprecipitation (lanes 1 and 5), a 200-kDa protein is detected with serum EC (lane 1, square dot). This band is absent when immunoprecipitation is performed with an irrelevant antiserum (lane 2), when extracts from nontransfected HeLa cells are used (lanes 3 and 4), or when the Western blot is probed with IC antiserum (lane 5). From these experiments we conclude that, as for the Notch2 receptor (7), no full-length Notch1 (p300) is expressed at the cell surface. Only associated processing products, which form a heterodimeric molecule able to bind the ligand, are expressed at the cell surface.
Figure 1.
(A) The p120 processing product of Notch1, but not the full-length p300 precursor, is present at the cell surface. Jurkat T cells were labeled for 2 hr with [35S]methionine and incubated with sulfo-NHS-LS biotin (+) or mock-treated (−). After lysis, extracts were immunoprecipitated with an antiserum raised against the intracellular region of Notch1 (antiserum IC) either directly (T) or after adsorption on streptavidin-agarose (B), and run on a 6.5% SDS-polyacrylamide gel. Positions of full-length Notch (p300) and p120 processing products are indicated. (B) Notch1 p120 is associated with the p200 N-terminal region of the molecule. Extracts from untransfected HeLa cells (lanes 3 and 4) or from HeLa cells stably transfected with murine Notch1 cDNA (lanes 1, 2, 5, and 6) were immunoprecipitated with the antiserum IC described above (lanes 1, 3, and 5) or with an irrelevant serum (lanes 2, 4, and 6) in TNT buffer. The precipitates were resolved on a 6.5% SDS-polyacrylamide gel, transferred onto Immobilon membranes, and probed with an antiserum raised against amino acids 1621–1637, located in the extracellular region (EC: lanes 1–4), or with serum IC (lanes 5 and 6). Molecular mass markers (kDa) are indicated on the left and the positions of p300, p200 (also marked by a square dot on the left), and p120 are shown on the right. ns, nonspecific band.
Inhibition of Notch Processing.
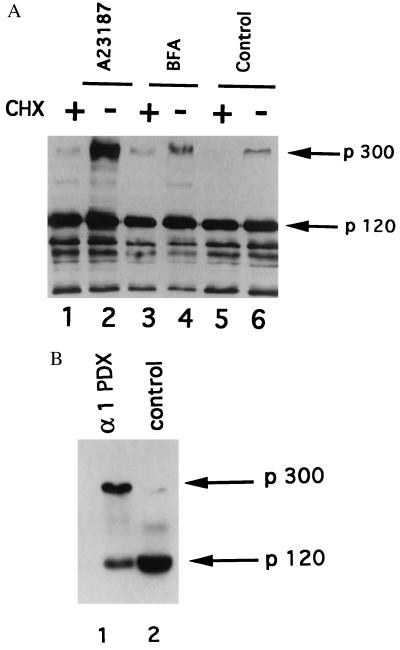
To characterize the subcellular compartment where Notch processing takes place, HeLa cells stably transfected with mouse Notch1 were treated with brefeldin A (BFA), a drug known to block the export from the endoplasmic reticulum (ER) to the Golgi complex (13), and with the calcium ionophore A23187, an inhibitor of calcium-dependent proteases of the secretory pathway (14). After treatment with BFA the processing of Notch1 was reduced (Fig. 2A, lane 4) when compared with the control cells (lane 5), as observed previously for Notch2 (7). In addition, when cells were treated with the calcium ionophore A23187 an important accumulation of p300 with no further processing into p120 could be observed (lane 2). The same experiments were carried out in the presence of cycloheximide (CHX) (Fig. 2A, lanes 1, 3, and 5). The results suggested that p300 is quite unstable whereas p120 is very stable. Therefore, we did not see any decrease in the amount of p120 when p300 processing was inhibited (lanes 2 and 4). It has been shown previously that a Notch1-derived construct starting at amino acid 1447 (plasmid LNG) was constitutively processed on the carboxyl side of the sequence RQRR (amino acids 1651–1654) after transfection into mouse fibroblasts (3). Interestingly, similar sequences can be found at similar positions in other species as well as in other Notch paralogs (not shown). This sequence is highly reminiscent of the consensus processing site [RX(K/R)R] of furin, a ubiquitously expressed membrane-associated, calcium-dependent endoprotease that processes proproteins in the constitutive secretory pathway (10). This is consistent with the effect of BFA and A23187 (Fig. 2A). To test the possible involvement of furin or a related family member, we examined the effect of the expression of α1-antitrypsin Portland (known as α1-PDX), a mutant of a natural elastase inhibitor that is a potent inhibitor of this family of convertases (15). α1-PDX has been shown to inhibit all known convertases in vitro (16) but only the intracellular convertases enriched within the constitutive secretory pathway in vivo (17). We generated Jurkat cells stably transfected with α1-PDX (the expression of α1-PDX was verified by immunofluorescence and Western blot analysis; not shown). Immunoblot analysis of one of these stable clones, C12D, indicated that Notch processing was strongly inhibited when compared with control cells (Fig. 2B).
Figure 2.
Inhibition of Notch1 receptor processing in the presence of BFA or A23187 in HeLa(N1) (A) and in the presence of α1-PDX in Jurkat cells (B). Identical amounts (100 μg) of the following extracts were run on a 6.5% SDS-polyacrylamide gel and incubated after transfer with antiserum IC. (A) Extracts from HeLa(N1) cells treated (3 hr) with BFA (50 μg/ml) (lanes 3 and 4) or with A23187 (2 μM) (lanes 1 and 2) in the presence (lanes 1, 3, and 5) or absence (lanes 2, 4, and 6) of 50 μg/ml cycloheximide. (B) Extracts from Jurkat cells (lane 2) or Jurkat cells stably transfected with the α1-PDX cDNA (clone C12D, lane 1). Positions of full-length Notch (p300) and p120 processing product are indicated.
Furin-Dependent Processing of the Notch1 Receptor.
To directly test the implication of furin itself in this process, we analyzed the biosynthesis and processing of Notch1 in LoVo cells, a cell line established from a human colon carcinoma that expresses no functional furin as a result of a mutation in the coding region (18). Fig. 3 shows the results of pulse–chase experiments carried out in LoVo or HeLa cells transiently transfected with Notch1: after 90 min of chase, p120 accumulated in HeLa cells (lanes 1–4), whereas it could not be detected in LoVo cells (lanes 5–8: the decrease of p300 in LoVo cells is a result of the intrinsic short half-life of this molecule; see Fig. 2). However, cotransfection of the furin cDNA into LoVo cells resulted in the reappearance of p120 (lanes 9–12), with a kinetics similar to that observed in HeLa cells. These results support the hypothesis that Notch is constitutively processed by furin.
In Vitro Cleavage of Notch1 Receptor by Purified Furin.
To precisely localize the site of cleavage of Notch1 by furin, we generated a recombinant Notch fragment encoding amino acids 1564–1723 (from the end of the third LNG repeat to the last amino acid N-terminal to the transmembrane region), fused to GST at its N terminus and to 6 histidines at its C terminus (Fig. 4A). This fragment includes, in particular, the putative furin site mentioned above. After purification on glutathione-agarose, the fusion protein was used as a substrate for in vitro reactions with purified recombinant furin. As shown in Fig. 4B in vitro cleavage of the fusion protein (45 kDa, lane 3, upper round dot) induced the appearance of 37- and 8-kDa fragments (lane 2; the glutathione S-transferase protein alone was not a substrate for furin, not shown). We purified the 8-kDa (His)6-tagged fragment on a nickel-agarose column (lane 1, arrow) and determined its amino-terminal sequence. The resulting sequence, ELDPM, indicates that furin indeed cleaves Notch1 between R1654 and E1655 at the physiologically relevant RQRR processing site for furin-like proteases.
Figure 4.
Mutational analysis of Notch digestion by furin in vitro. (A) Schematic map of the recombinant substrate: murine Notch1 amino acids coordinates are indicated below the map. The main processing site (RQRR) and the two potential secondary sites are shown above the map, and the positions where processing by furin takes place are shown below. (B) The wild-type (WT, lanes 2 and 3) or the mutated (RQRR → AAAA; Mut, lanes 4 and 5) recombinant substrates were either mock-digested (lanes 3 and 4) or digested with purified recombinant BCRD-furin (lanes 2 and 5), and the products were analyzed on 15% Tricine gels followed by silver staining. The full-length substrate and N-terminal digestion products are indicated by dots on the left, and the C-terminal digestion products are indicated by arrows. The digestion product, purified on nickel-agarose, used for N-terminal sequencing is shown in lane 1. Molecular mass markers (kDa) are indicated on the right.
Mutagenesis of the Furin Cleavage Site.
To test the in vivo effect of mutating the furin cleavage site, we introduced the RQRR → AAAA mutation into the full-length Notch1 cDNA, which then was stably transfected into HeLa cells or transiently into 293T cells. Western blot analysis of transfected 293T cells showed that when compared with the wild-type form, processing of the mutated Notch1 was almost completely abolished (Fig. 5, compare lane 3 with lane 2). Besides, immunofluorescence analysis of HeLa cells stably transfected with such a mutated Notch1 cDNA demonstrated its accumulation in the ER and Golgi compartments whereas little or no plasma membrane expression could be observed (not shown). However, some p120 could still be detected, and this amount increased when a cDNA encoding furin was cotransfected (Fig. 5, lanes 3 and 7). To test the possibility that the observed cleavage might be because of processing by furin at potential secondary sites (KR: 1632–1633) and/or (RK: 1627–1628), we synthesized a recombinant substrate similar to the one described above but carrying the RQRR → AAAA mutation in the principal site. As shown in Fig. 4B (lanes 4 and 5), digestion of this substrate by furin generated a 10-kDa peptide, which is probably the product of cleavage at one of the two dibasic sites. Therefore, the band observed in Fig. 5 (lanes 3 and 7) is likely to be a result of processing at one of these two secondary sites (the migration difference because of the generation of a molecule larger than p120 by about 20 aa would not be detectable). To test this hypothesis, we mutated the three sites and introduced the resulting Notch1 cDNA into 293T cells: under these conditions no p120 could be detected, either in the absence (lanes 4 and 5) or in the presence (lanes 8 and 9) of cotransfected furin cDNA, confirming the involvement of furin in the constitutive processing of Notch.
Figure 5.
Processing of mutated Notch molecules transfected into 293T cells. Extracts from 293T cells either untransfected (lane 1) or transfected with full-length Notch1 cDNA (wt, lanes 2 and 6), mutated Notch1 cDNA (m1, mutation RQRR → AAAA; lanes 3 and 7), and Notch1 cDNA carrying the three mutations described in the text (m3 and m′3: lanes 4 and 5 and 8 and 9) in the absence (lanes 2–5) or presence (lanes 6–9) of cotransfected furin cDNA were analyzed by immunoblotting by using the IC antiserum. Positions of full-length Notch (p300) and p120 processing products are indicated. m3 and m′3 correspond to two independent experiments by using the Notch1 triple mutant.
DISCUSSION
The proteolytic activity responsible for p300 processing plays a critical role in Notch signaling as it determines the functional structure of the receptor. Our data demonstrate that Notch1 is present at the cell surface as a heterodimeric molecule (p120/p200), whereas the precursor protein (p300) does not reach the cell surface and probably is cleaved into p120 and p200 in the trans-Golgi Network (TGN). We further demonstrate that a furin-like convertase is responsible for this constitutive processing, which takes place C-terminal to the RQRR sequence (amino acids 1651–1654). Mutation of the furin cleavage sites produces full-length, 300-kDa molecules that are not processed and do not reach the cell surface. Proteolytic maturation also is absent in LoVo cells that are devoid of furin activity. Furin is known to cycle between the TGN and the plasma membrane, consistent with the localization of the processing step presented above. Furin belongs to the subtilisin/kexin family of proprotein convertases. Additional members of this family include PC1, PC2, PACE4, PC5/6, and PC7 (10, 19). Although these proteins exhibit varying tissue-specific distributions and have similar cleavage recognition sequences, only furin exhibits a ubiquitous tissue expression. Because the Jurkat cells where we observe processing only express furin, PC5/6, and PC7 (16), and because LoVo cells, where Notch1 processing does not take place, express PACE4 and PC7, but not PC5/6 (10, 19), Notch might alternatively be processed by PC5/6. But because furin is the only ubiquitously expressed member of this family, and until a more detailed tissue analysis is performed, we propose that furin is the major candidate enzyme responsible for the constitutive processing of Notch.
Recently it has been suggested that the disintegrin metalloprotease KUZ encoded by the gene Kuzbanian is required for the constitutive cleavage of Notch (8). In particular, the authors show that overexpression of a dominant negative mutant of Drosophila KUZ (KUZ-DN) lacking the pro- and metalloprotease domains results in no detectable p120 both in transfected S2 cells and in Drosophila imaginal discs. The authors also show that no processing of Notch can be observed in kuz null Drosophila embryos. In our experiments, the expression of KUZ-DN did not affect mouse Notch1 processing, irrespective of the amount of this dominant negative molecule we introduced into cells (data not shown). In any case, further work will be necessary to clarify these apparent discrepancies and determine whether they are because of differences in the experimental systems used, for example, between Drosophila and mammals.
Because KUZ is required for the lateral inhibition process during Drosophila neurogenesis (9) and its target seems to be the extracellular region of Notch (20, 21), we think that KUZ is not involved in the constitutive maturation of the receptor but in a subsequent processing step that would follow interaction with the ligand. This might explain the recently published observation that a new band migrating slightly faster than p120 is induced by contact between cells expressing Notch1 and cells expressing Jagged2 (22). That membrane metalloproteases of the ADAM family are postulated to act at the cell surface (23) is in favor of this hypothesis. Indeed results from our lab (S.J., O.L., E. Hirsinger, O. Pourquié, B. Vaznum-Finney, F.L., C.B., N.G.S., and A.I., unpublished data) have shown that KUZ-DN is able to inhibit transactivation of a target gene of the Notch pathway induced by ligand binding to the receptor. In conclusion, mammalian Notch molecules are constitutively processed by a furin-like convertase as part of their normal maturation process, and this processing is required for cell surface expression of a heterodimeric functional receptor.
Acknowledgments
We thank F. Schweisguth (Ecole Normale Supérieure, Paris) and S. Whiteside for critical reading of the manuscript, D. Pan and G. M. Rubin (Univ. of Berkeley, CA) for the kind gift of the kuz-DN construct, N. Rice (National Cancer Institute, Frederick Cancer Research and Developmental Center, Frederick, MD) for the choice of the peptide used for raising the EC serum, and J. S. Munzer (Clinical Research Institute of Montreal, Montreal, Canada) for preparing BCRD-furin. This research was sponsored in part by grants from Association pour la Recherche sur le Cancer, Agence Nationale de Recherches sur le SIDA, Institut National de la Santé et de la Recherche Médicale, and Ligue Nationale Francaise contre le Cancer to A.I. and a group grant from the Medical Research Council of Canada to N.G.S.
ABBREVIATION
- BFA
brefeldin A
References
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Fleming R J, Purcell K, Artavanis-Tsakonas S. Trends Cell Biol. 1997;7:437–441. doi: 10.1016/S0962-8924(97)01161-6. [DOI] [PubMed] [Google Scholar]
- 3.Kopan R, Schroeter E H, Nye J S, Weintraub H. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarriault S, Brou C, Logeat F, Schroeter E, Kopan R, Israel A. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 5.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 6.Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Blaumueller C M, Qi H L, Zagouras P, Artavanis-Tsakonas S. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 8.Pan D J, Rubin G M. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 9.Rooke J, Pan D, Xu T, Rubin G M. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 10.Seidah N G, Chretien M, Day R. Biochimie. 1994;76:197–209. doi: 10.1016/0300-9084(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 11.Rebay I, Fleming R J, Fehon R G, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 12.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 13.Klausner R D, Donaldson J G, Lippincott-Schwartz J. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klenk H D, Garten W, Rott R. EMBO J. 1984;3:2911–2915. doi: 10.1002/j.1460-2075.1984.tb02231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson E D, Thomas L, Hayflick J S, Thomas G. J Biol Chem. 1993;268:24887–24891. [PubMed] [Google Scholar]
- 16.Decroly E, Wouters S, Di Bello C, Lazure C, Ruysschaert J M, Seidah N G. J Biol Chem. 1996;271:30442–30450. doi: 10.1074/jbc.271.48.30442. [DOI] [PubMed] [Google Scholar]
- 17.Benjannet S, Savaria D, Laslop A, Munzer J S, Chretien M, Marcinkiewicz M, Seidah N G. J Biol Chem. 1997;272:26210–26218. doi: 10.1074/jbc.272.42.26210. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi S, Kasai K, Hatsuzawa K, Kitamura N, Misumi Y, Ikehara Y, Murakami K, Nakayama K. Biochem Biophys Res Commun. 1993;195:1019–1026. doi: 10.1006/bbrc.1993.2146. [DOI] [PubMed] [Google Scholar]
- 19.Seidah N G, Hamelin J, Mamarbachi M, Dong W, Tardos H, Mbikay M, Chretien M, Day R. Proc Natl Acad Sci USA. 1996;93:3388–3393. doi: 10.1073/pnas.93.8.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sotillos S, Roch F, Campuzano S. Development. 1997;124:4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- 21.Wen C, Metzstein M M, Greenwald I. Development. 1997;124:4759–4767. doi: 10.1242/dev.124.23.4759. [DOI] [PubMed] [Google Scholar]
- 22.Luo B, Aster J C, Hasserjian R P, Kuo F, Sklar J. Mol Cell Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfsberg T G, White J M. Dev Biol. 1996;180:389–401. doi: 10.1006/dbio.1996.0313. [DOI] [PubMed] [Google Scholar]