Abstract
The immortalization of human cells is a critical step in multistep carcinogenesis. Oral-esophageal carcinomas, a model system to investigate molecular mechanisms underlying squamous carcinogenesis, frequently involve cyclin D1 overexpression and inactivation of the p53 tumor suppressor. Therefore, our goal was to establish the functional role of cyclin D1 overexpression and p53 inactivation in the immortalization of primary human oral squamous epithelial cells (keratinocytes) as an important step toward malignant transformation. Cyclin D1 overexpression alone was found to induce extension of the replicative life span of normal oral keratinocytes, whereas the combination of cyclin D1 overexpression and p53 inactivation led to their immortalization. This study also demonstrates that immortalization of oral keratinocytes can be independent of telomerase activation, involving an alternative pathway of telomere maintenance (ALT).
Introduction
Oral-esophageal squamous epithelial cells share major structural and functional features with the well-characterized skin keratinocyte systems and provide a good model to study basic keratinocyte biology as well as processes of immortalization and malignant transformation. Keratinocytes undergo an exquisite program of proliferation and differentiation in vivo. Insights into underlying molecular mechanisms in oral-esophageal keratinocytes have been gained through in vitro and in vivo models (1, 2).
Normal or primary keratinocytes display a restricted replicative life span in cell culture. Those cells initially proliferate but eventually enter a state of permanent growth arrest, called replicative senescence (3), which is clearly distinct from terminal differentiation. Senescence is accompanied by several genetic changes including an increase in expression of several inhibitors of cyclin-dependent kinases (cdk’s), as well as telomere shortening (4, 5). It has been suggested that senescence forms a barrier against tumorigenesis and that acquisition of the ability to proliferate an unlimited number of times, termed immortalization, is an essential step in the malignant transformation of normal cells (6). Tumor cells escape this growth control checkpoint and therefore can provide some insight into the mechanisms involved in the pathway from senescence to immortality. Furthermore, some of the most common known genetic changes in cancer development, such as the inactivation of the p53 and pRb pathways, play a critical role in cellular immortalization (7). As a separate consideration, cell culture conditions may contribute to immortalization (8).
In addition to changes in oncogenes and tumor suppressor genes, the gradual loss of DNA from the ends of telomeres has been implicated in the control of the proliferative potential of cells (9). Shortened telomeres at a critical length are thought to provide the signal to activate the program of cellular senescence. Telomerase is an enzyme that restores telomeric DNA sequences, and expression of its activity is thought to be involved in immortalization of human cells in vitro and eventually tumor progression in vivo (10). Telomerase activity has been detected in the majority of different tumor types, including oral squamous cell carcinoma (11). Presenescent human cells lack telomerase, and ectopic expression of hTERT (the catalytic subunit of telomerase) in presenescent human fibroblasts or retinal pigment epithelial cells was found to immortalize these cell types (12). By contrast, it has been shown that ectopic expression of hTERT is not sufficient to immortalize normal human epithelial cells and that additional loss of the pRb/p16(INK4a) cell cycle control is required (13, 14). Generally, one can also bypass senescence through inactivation of the pRb and p53 tumor suppressor pathways, e.g., by SV 40 large T antigen (15). Such cells remain telomerase negative and eventually enter crisis characterized by cell death. Activation of telomerase will permit such cells with critically short telomeres to become immortal. Therefore, the most common method to generate immortalized cells is with oncogenic viruses such as SV 40 or human papillomavirus (HPV) (16, 17). Recently, it has been demonstrated that telomerase can cooperate with SV 40 T antigen and ras to induce malignant transformation in normal human cells (6, 18).
Overexpression of the cyclin D1 oncogene is associated with human cancer and is the most common genetic alteration in human oral-esophageal squamous cell carcinomas (19, 20). Furthermore, cyclin D1 induces dysplasia in the oral-esophageal epithelia of transgenic mice (21). Cyclin D1 associates with cdk 4 and 6, and the complex phosphorylates and inactivates pRb; inactivating mutations of pRb are not observed in oral-esophageal squamous cell carcinomas. By contrast, p53 mutations are frequently found in oral-esophageal cancer. Furthermore, p53 is inactivated in a high proportion of oral dysplastic lesions, implicating a role in the initiation of immortalization in this cell type (22). Following these patterns of genetic events seen during in vivo tumor development, we sought to identify the role of cyclin D1 overexpression and functional p53 inactivation in the immortalization of human oral squamous epithelial cells or keratinocytes. Cyclin D1 alone or in combination with dominant-negative p53 was ectopically overexpressed in normal human oral keratinocytes in culture using retroviral transduction. Whereas cyclin D1 overexpression extended the replicative life span of primary human oral keratinocytes, the combination with dominant-negative p53 resulted in immortalization of these cells. Importantly, immortalization under these conditions was independent of telomerase activity.
Methods
Cell culture and retroviral infection.
Normal diploid human oral keratinocytes (OKF6) were established from a biopsy of normal floor of the mouth of clinically and genetically normal tissue. OKF6 cells have been cryopreserved within their first two serial passages in culture and characterized extensively (23). OKF6 and the lines generated thereof (OKF6-LacZ, OKF6-D1, OKF6-Δp53, OKF6-D1/Δp53) were grown in defined keratinocyte-SFM with defined growth supplement (Life Technologies Inc., Rockville, Maryland, USA) and final Ca2+-concentration of 0.4 mM. The medium was supplemented with antibiotics. To assess mitogen dependence, cells were plated in defined keratinocyte-SFM supplemented with a reduced concentration (10%) of growth factors. The amphotropic packaging cell line Phoenix A was grown in supplemented DMEM (Sigma Chemical Co., St. Louis, Missouri, USA) and then transiently transfected with the respective retroviral vector to generate amphotropic retroviruses. Retroviral supernatant was harvested 48 hours after transfection and filtered through a 0.45-μm filter, and fresh retroviral supernatant was used for infection of exponentially growing OKF6 cells. Medium containing puromycin (1 μg/ml) and/or G418 (400 mg/ml) was exchanged to start selection 48 hours after infection. In each set of experiments, multiple clones were pooled for further processing after 1–2 days of puromycin selection and/or 5 days of G418 selection. The retroviral expression vectors pBPSTR-D1 and pBABE-LacZ were obtained from S.A. Reeves (Massachusetts General Hospital, Boston, Massachusetts, USA) (24). The inducible retroviral vector pBPSTR-D1 contains both elements of the tetracycline-regulated system and has been described previously by us and others (25). The gene of interest is expressed in the absence of tetracycline (TET-OFF vector system). The puromycin resistance gene under the control of the promoter within the 5′-LTR is present in both retroviral vectors. The LXSN vector containing the p53 coding sequence with a V143A mutation was used to generate a dominant-negative version of p53 (26).
Determination of replicative life span.
Serial cultures of the different OKF6 lines (OKF6, OKF6-LacZ, OKF6-D1, OKF6-Δp53, OKF6-D1/Δp53) were performed in 10-cm dishes by plating 105 cells, refeeding the cells every 2nd day, and subculturing every 4–5 days. The doubling number of each passage was calculated using the formula PD = (nf/n0)/log2, where n0 is the initial number of cells and nf is the final number of cells. Cellular immortalization was defined as cell growth of at least three times beyond the life span of the parental cells.
Flow cytometry.
Exponentially growing cells were collected at different population doubling (PD) times and centrifuged at 300 g for 4 minutes. The cell pellet was resuspended in 0.5 ml of PBS, fixed in 5 ml 70% ethanol, and stored at –20°C overnight. The cells were then washed twice with PBS and resuspended in a 1-ml solution containing 3.8 mM sodium citrate and 10 μg/ml propidium iodide. After 10 mg/ml of RNase treatment at 37°C for 20 minutes, the samples were analyzed by a fluorescence-activated cell sorter (FACScan; Becton Dickinson & Co., Franklin Lakes, New Jersey, USA).
Western blot analysis.
Lysates from exponentially growing cells were harvested in a buffer (50 mM HEPES [pH 7.4], 0.1% Nonidet P-40, and 250 mM NaCl) with 1 mM protease and 10 mM phosphatase inhibitors. A total of 10 μg of total protein was separated on 10% SDS-polyacrylamide gels and transferred to Immobilon membranes (Millipore Corp., Bedford, Massachusetts, USA). Blocking was performed in 5% milk, 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.2% Tween-20 for 1 hour, followed by incubation with primary antibodies as indicated (1:3,000). The secondary antibody was either peroxidase-conjugated anti-mouse or anti-rabbit Ig (1:2,500; Amersham Corp., Burlington, Massachusetts, USA). Detection was by chemiluminescence (ECL; Amersham Corp.). Primary antibodies used were monoclonal cyclin D1 antibody (HD11), polyclonal cyclin D1 antibody (H295), monoclonal p53 antibody 421, monoclonal p53 antibody 122, and monoclonal p16 antibody (JC8). Quantification was done using Image software (National Institutes of Health, Bethesda, Maryland, USA).
TRAP assays/telomeric-length assays.
Cellular extracts of all generated oral keratinocytes were assayed for telomerase activity using the PCR-based telomerase repeat amplification protocol (TRAP) assay (27). Cellular extracts (50 and 500 ng), along with a heat-inactivated controls, were used for TRAP assays. Telomere length was measured by hybridizing a 32P-labeled telomeric (CCCTAA) probe to 10 μg of HinfI- and RsaI-digested genomic DNA as described previously (27).
Karyotypic analysis.
Exponentially growing cultured OKF6 and OKF6-D1/Δp53 cells were incubated with 10–7 M Colcemid (Life Technologies Inc.) for 1 hour, resuspended in warm hypotonic buffer (0.075 M KCl), allowed to swell for 10–15 minutes at 37°C, fixed by addition of cold ethanol/acetic acid, and applied to chilled microscope slides. Chromosome preparations of cultured cells were counted for the number of metaphase spreads. Metaphase spreads (> 10) were then analyzed at different PD times at the University of Pennsylvania Cytogenetics Core Facility.
Subcutaneous tumorigenicity assays.
Six- to eight-week-old immunocompromised athymic nude mice (NIH III; Charles River Laboratories, Wilmington, Massachusetts, USA) were injected with a cell suspension containing 2 × 106 exponentially growing cultured OKF6, OKF6-D1/Δp53, SCC15 (oral squamous cancer cell line), or TE 12 (esophageal squamous cancer cell line) cells. Tumor size was measured every 3 days. The time of initial tumor formation was defined as the time when the tumor had reached a diameter of 3 mm. Mice were sacrificed when the tumors grew to 1 cm in diameter or after 12 weeks of monitoring and were subsequently analyzed.
Results
Ectopic cyclin D1 overexpression in normal human oral keratinocytes induces cell-cycle abnormalities and extension of replicative life span.
To elucidate the role of the cyclin D1 oncogene in the immortalization of oral squamous epithelial cells, we ectopically overexpressed cyclin D1 in normal human oral keratinocytes. To overexpress cyclin D1, we used a tetracycline-regulated retroviral vector system, pBPSTR-D1, containing the human cyclin D1 cDNA, to infect the normal human oral keratinocytes (OKF6). OKF6 cells have been described previously and shown not to bear any genetic alterations (14, 23).
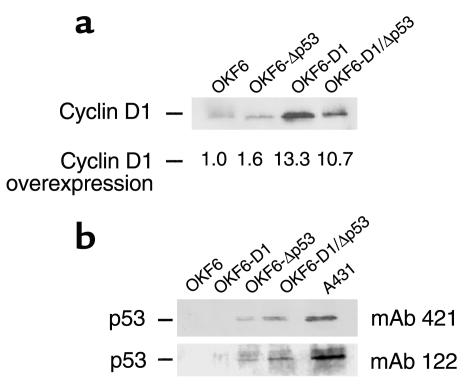
A tetracycline-inducible system was used for infection in order to prevent any potential toxic effects of cyclin D1 overexpression. Given that no toxicity was observed, cyclin D1 was constitutively expressed (TET off) in the ensuing experiments (25). Infection efficiency was monitored and confirmed with a lacZ gene containing retrovirus (pBabe-lacZ). Infected OKF6 cells were selected with the appropriate antibiotic, and the selected clones were pooled to ensure polyclonality. Cells used for further studies were designated OKF6-D1. The levels of endogenous and ectopically expressed cyclin D1 in parental OKF6 and cyclin D1–infected cells (OKF6-D1 cells) were determined using Western blot analysis (Figure 1a).
Figure 1.
Effects of cyclin D1 overexpression and/or dominant-negative p53 expression in oral keratinocytes (OKF6). (a) Equal amounts of protein for parental OKF6, OKF6-Δp53, OKF6-D1, and OKF6-D1/Δp53 were separated on a 10% SDS-PAGE. The level of cyclin D1 overexpression was compared by subsequent immunoblot analysis using polyclonal cyclin D1 antibody (H295) and monoclonal cyclin D1 antibody (HD11) (data not shown). (b) Equal amounts of protein for parental OKF6, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 were separated on a 10% SDS-PAGE. Subsequent immunoblot was done with two different conformational p53 antibodies: the monoclonal p53 antibody 421 and the monoclonal p53 antibody 122. The human epidermoid cancer cells A431 served as positive control.
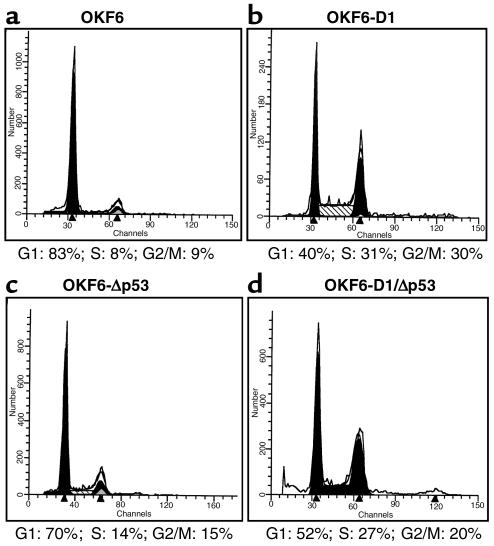
Because cyclin D1 plays an important role in the G1-phase progression of the cell cycle, we investigated the effects of cyclin D1 overexpression on cell-cycle dynamics in these cells. Flow cytometry analysis revealed pronounced cell-cycle redistribution in OKF6-D1 cells, with a 20% increase in cells cycling in S-phase compared with parental OKF6 cells (Figure 2, a and b). Similar results were observed in five separate cyclin D1 transduction experiments (data not shown).
Figure 2.
Cell-cycle pattern of the parental and derived oral keratinocytes. Cell-cycle analysis of OKF6, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 cells was done under standard conditions using FACScan. Cell-cycle analysis is displayed either as a histogram or as a table. (a) OKF6; (b) OKF6-D1; (c) OKF6-Δp53; and (d) OKF6-D1/Δp53.
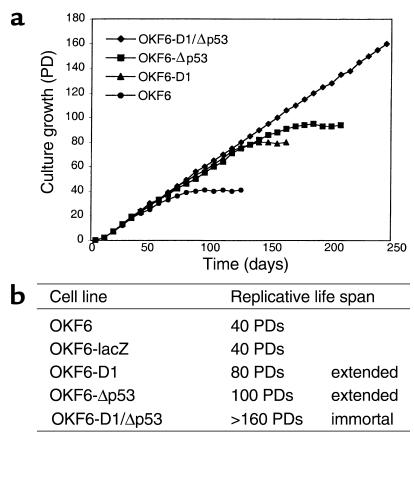
We next investigated the growth of these cells on extended passage in culture. Both OKF6 cells and OKF6 cells transduced with the control vector (OKF6-LacZ) senesced at 40 PDs, as evidenced by growth arrest and enlargement of cells in culture (Figure 3). By contrast, the replicative life span of OKF6-D1 cells was significantly and reproducibly extended to 80 PDs, at which time the OKF6-D1 cells eventually senesced (Figure 3). Of note, the p16 tumor suppressor protein product was downregulated in early passages, but expression was high in presenescent passages of OKF6-D1 cells (data not shown).
Figure 3.
Replicative life span of the parental and derived oral keratinocytes. (a) The growth characteristics of the parental and derived OKF6 cells are depicted as follows: OKF6 (circles), OKF6-D1 (triangles), OKF6-Δp53 (squares), and OKF6-D1/Δp53 (diamonds). (b) The replicative life span of OKF6, OKF6-lacZ, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 was assessed by calculating the PDs of each cell line. Immortalization was assessed if cells grew at least three times beyond the life span of the parental cells.
Ectopic expression of dominant-negative p53 in OKF6-D1 cells results in immortalization.
Given that inactivation of p53 appears to be important in the early phases of oral-esophageal carcinogenesis, we investigated the functional consequences of p53 inactivation in combination with cyclin D1 overexpression in human oral keratinocytes. Independent transduction experiments were done to express a dominant-negative version of the tumor suppressor gene p53 (V143A) in OKF6-D1 cells. Clones from each experiment were pooled and established as OKF6-D1/Δp53 oral keratinocytes. To exclude the possibility that changes occurred as a consequence of specific genetic selection in one generated cell strain, a second strain from a different transduction experiment was established. Both strains revealed identical genetic features and growth properties.
The effect of the dominant-negative p53 mutant was demonstrated by stabilization of the p53 protein in infected cells as assayed by Western blot analysis using two different conformational p53 antibodies (Figure 1b). Both strains of OKF6-D1/Δp53 cells revealed stabilized p53 protein as did OKF6-Δp53 cells compared with OKF6-D1 cells or the parental oral keratinocytes, where no p53 signal was detected (Figure 1b). FACS analysis of OKF6-D1/Δp53 cells revealed a cell-cycle redistribution comparable to that of the OKF6-D1 cells, whereas the OKF6-Δp53 cells showed no significant cell-cycle redistribution (Figure 2, c and d). The FACS analysis was done at different PD times (early PD and late PD), but no major differences were observed (data not shown).
The OKF6-D1/Δp53 cell strains revealed an extension of their replicative life span to more than 160 PDs and continue to grow in culture (Figure 3). Thus, overexpression of both cyclin D1 and dominant-negative p53 reproducibly immortalizes human oral keratinocytes. It is conceivable that indefinite passage in culture will eventually result in the acquisition of other genetic alterations and frank transformation. We did not detect the onset of any widespread cell death while passaging the OKF6-D1/Δp53 cell strains as observed during crisis in SV 40 immortalized cells. Apoptosis was not observed by light microscopic evaluation or by DNA-laddering (Figure 3 and data not shown) in spite of a small sub-G1 peak in the FACS analysis. Thus, the sub-G1 peak observed in the OKF6-D1/Δp53 cells does not appear to be consistent with apoptosis.
OKF6-Δp53 cells showed an extension of the replicative life span (100 PDs), which was slightly more pronounced than that in the OKF6-D1 cells, but they ultimately senesced (Figure 3). Interestingly, there was only a minimal increase in the polyploid cell population in OKF6-Δp53 cells during serial passage (data not shown). Although p16 was downregulated in early-passage OKF6-D1 cells and then expressed in presenescent cells, it was continuously expressed in OKF6-D1/Δp53 cells (data not shown).
Mechanism of immortalization in OKF6-D1/Δp53 cells.
Immortal human cells maintain their telomeres by one of two known mechanisms: activation of telomerase or activation of an alternative pathway in lengthening telomeres (ALT).
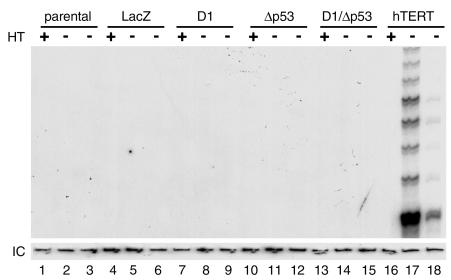
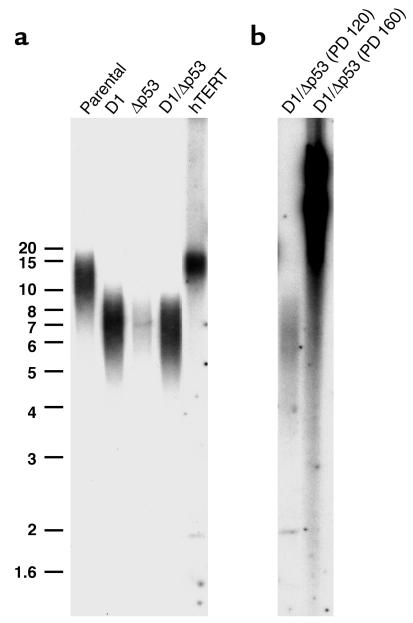
To further investigate the mechanisms of immortalization in oral keratinocytes, we assayed the parental OKF6 cells and derivatives for telomerase activity. Telomerase activity was absent in all OKF6 cells (OKF6, OKF6-LacZ, OKF6-D1, OKF6-Δp53, OKF6-D1/Δp53), including the immortal OKF6-D1/Δp53 cell strains (Figure 4). Telomere length measurements by Southern blotting with a probe specific for mammalian telomeric repeats (TRF) revealed a progressive shortening of telomeres that correlated with the replicative life-span extension of oral keratinocytes. Parental OKF6 cells showed a longer average telomere length than did OKF6-D1 and OKF6-Δp53 cells, which retained longer telomeres than OKF6-D1/Δp53 cells at 120 PDs (Figure 5). Interestingly, the OKF6-D1/Δp53 cells at 160 PDs revealed an elongation of telomere length as measured by TRF with a heterogeneous lengthening including very long telomeres of up to 40 kb (Figure 5). Thus, the OKF6-D1/Δp53 cell strains acquired the ability to maintain their telomeres in the absence of telomerase activity at PD 120–160.
Figure 4.
Telomerase activity in the parental and derived oral keratinocytes. Cellular extracts (500 ng and 50 ng) of OKF6, OKF6-lacZ, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 cells were assayed for telomerase activity using the PCR-based TRAP-assay. Heat-treated (HT)samples served as negative control, and OKF6 cells transduced to express hTERT (13) were used as positive control. IC is an internal PCR-control to demonstrate the absence of PCR inhibitors in the cellular extracts.
Figure 5.
Telomere length in the parental and derived oral keratinocytes. (a) Telomere length for OKF6, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 (120 PDs) cells was analyzed by hybridization of genomic DNA with a specific oligonucleotide probe. OKF6-hTERT cells served as control. (b) Telomere length for OKF6-D1/Δp53 (120 PDs) and OKF6-D1/Δp53 (160 PDs) cells. Size standard is indicated at the left. The broad smear in OKF6-D1/Δp53 (160 PDs) starts at a telomere length of 40 kb.
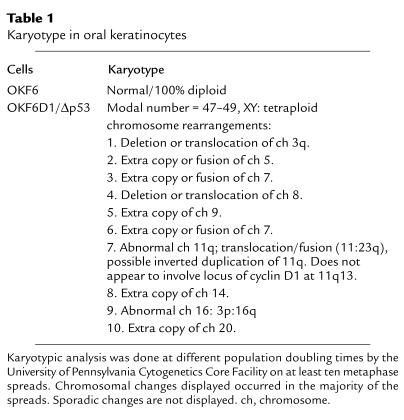
To assess for chromosomal recombination events in the OKF6-D1/Δp53 cells, karyotypic analysis was performed at different PD times. This revealed aneuploidy and consistent chromosomal rearrangements compatible with the selection of chromosomal changes in cells with critically short telomeres (Table 1).
Table 1.
Karyotype in oral keratinocytes
Growth characteristics of immortalized OKF6-D1/Δp53 cells.
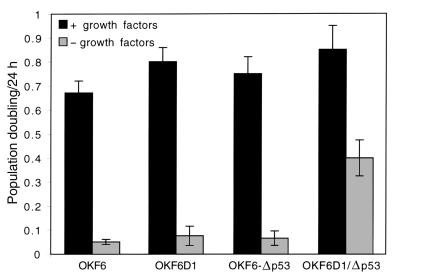
Next, we determined whether the process of immortalization in OKF6-D1/Δp53 cells impaired specific keratinocyte growth control mechanisms. We investigated the ability of OKF6, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 cells to grow in growth factor–reduced medium. Nonimmortalized oral keratinocytes (OKF6, OKF6-D1, OKF6-Δp53) completely retained dependence on growth factors (Figure 6) and did not grow in growth factor–reduced medium. By contrast, the immortalized OKF6-D1/Δp53 cell strains displayed a proliferative capacity, albeit reduced to 40% (Figure 6).
Figure 6.
Growth factor dependence of the parental and derived oral keratinocytes. OKF6, OKF6-D1, OKF6-Δp53, and OKF6-D1/Δp53 cells were plated at low density either in growth factor–supplemented (+ growth factors) or reduced (– growth factors) medium. Cells were counted 7–9 days later, and growth under these conditions was calculated as PDs per day. Results shown are an average of three experiments.
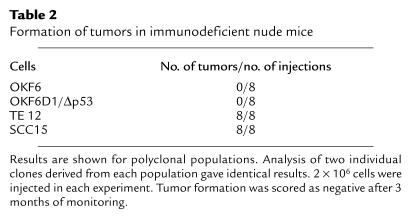
To assess whether the immortalized oral keratinocytes might have undergone malignant transformation, we injected the OKF6-D1/Δp53 cell strains into immunodeficient nude mice. As with OKF6 cells serving as negative controls, no tumors were observed with the OKF6-D1/Δp53 cell strains after 3 months of observation (Table 2). As positive controls, the human squamous epithelial cancer cell lines SCC15 (oral) and TE 12 (esophageal) readily formed tumors in this assay (Table 2).
Table 2.
Formation of tumors in immunodeficient nude mice
Discussion
One major difference between normal mammalian cells and cancer cells is their proliferative potential. Normal mammalian cells proliferate for a limited number of PDs and then enter a nondividing state known as senescence (3). Cancer cells are able to proliferate indefinitely and are therefore referred to as immortalized. Cellular immortalization is an essential step toward malignant transformation of normal cells. Important genetic changes associated with immortalization are loss of p53 and pRb functions, and activation of a telomere maintenance mechanism, typically telomerase (7).
That viral oncoproteins such as HPV16 E6 and E7, as well as SV 40 large T antigen, contribute to immortalization already indicates that both p53 and pRb pathways are involved (15, 16). Regardless of the precise mechanism by which the disruption of the p53 and pRb pathways induces immortalization, their importance is further supported by the well-known observation that these pathways are frequently altered in primary tumors and tumor-derived cell lines (28, 29). In this context, cyclin D1 overexpression or loss of p16/INK4A may be viewed as equivalent to loss of pRb function. Cyclin D1 overexpression is observed in different cancer types, especially of squamous origin, indicating a critical role of the cyclin D1 oncogene in carcinogenesis of this cell type (19, 20). Cyclin D1 overexpression is not only a frequent genetic event in the majority of oral cancers, but its expression is correlated with reduced disease-free and overall survival in this cancer (30). The oncogenic role of cyclin D1 was initially established in murine cells as cooperating with the ras, E1A, and neu oncogenes (31–33).
p53 is frequently mutated in oral squamous cell carcinomas. In oral premalignant lesions, expression of p53-positive cells in the suprabasal layers of the epithelium has been seen as an indication of impending malignant development. The prognostic significance of p53 gene mutations in these tumors has been demonstrated (34). That the loss of p53 function is directly associated with escape from senescence was initially demonstrated through the introduction of a dominant-negative p53 gene in fibroblasts (35). Dominant-negative p53 permits fibroblasts to proliferate for a limited number of PDs, beyond the point at which their normal counterparts become senescent. Evidence that this was attributable to loss of wild-type p53, and not gain of p53 function, was provided by studies with Li-Fraumeni fibroblasts (36).
Telomerase activity has been associated with many different tumor types. Up to 80% of primary head and neck squamous cell cancer specimens possessed telomerase activity, and its activity in oral rinses was proposed as a potential molecular marker (11). Several lines of evidence have implicated telomere erosion as the major checkpoint in limiting the proliferative potential of human cells (9). With each cell division, a portion of telomeric DNA is lost. Once the telomeres shorten to a critical length, the signal for the senescence program is activated. This telomere signal that activates the program of senescence could operate through the pRb and p53 pathways. Viral oncoproteins HPV E6 and E7 reverse the senescence program by inducing telomerase activation and targeting p53 and pRb (37). By contrast, expression of the catalytic subunit of telomerase independently was implicated in extension of the replicative life span in a subset of cells without involving pRb and p53 pathways (12). Other cells, including keratinocytes, appear to require at least additional loss of the pRb-mediated cell-cycle control mechanism as a major event toward immortalization (13, 14). Recently, it has been demonstrated that the nature of the cell culture conditions might also be an important factor in this cell type–specific process of immortalization (8).
We have demonstrated that OKF6 can extend their limited replicative life span by overexpressing the cyclin D1 oncogene. Combining this with inactivation of p53 leads to immortalization of the oral keratinocytes. Furthermore, the immortalized epithelial cells (OKF6-D1/Δp53) show reproducibly no increase in telomerase activity. Thus, the immortalization of oral keratinocytes induced by overexpression of cyclin D1 and inactivation of wild-type p53 appears to be independent of telomerase activation. pRb/p53 inactivation permitted continuous cell division beyond senescence and progressive telomere shortening. This result does not involve selection of cells with rare genetic alterations, which was confirmed through independent transduction experiments. In contrast to the results presented here, immortalization of epithelial cells using viral oncoproteins HPV E6 and E7 involves E6-dependent activation of telomerase (13).
Expression of SV 40 large T antigen in primary cells leads to inactivation of the pRb and p53 tumor suppressor pathways and allows such cells to grow beyond senescence. Such post-senescent cells lose telomeric sequences corresponding to continued cell division until they reach crisis, when telomeres are critically short and cells experience marked genetic instability (38). Surviving cells maintained telomere length through either activation of telomerase or utilization of ALT thought to involve a recombination based mechanism. Here we show that the combination of cyclin D1 overexpression and p53 inactivation leads to immortalization of oral keratinocytes without activation of telomerase. Telomeres in these immortalized cells are long and heterogeneous, suggesting that such cells maintain telomere length through ALT. Although most immortal cell lines and human tumors are believed to maintain telomere length through activation of telomerase, 10%–15% of tumors (39) and up to 40% of SV40-immortalized cell lines lack detectable telomerase (40). The mechanism of ALT in mammalian cells is not yet understood, whereas in yeast, ALT is accomplished through recombination involving RAD 52–mediated DNA recombination (41, 42).
The fact that telomere length, not telomerase activation, is the biochemical feature most closely associated with immortalization is illustrated by the telomerase-negative immortalized cells (39, 40) and is further supported by studies with telomerase dysfunctional mice (43). Although telomere biology may be different in mice, cells from late-generation telomerase-deficient mice are also forced to use alternative mechanisms capable of maintaining telomeres. Studies on mouse embryo fibroblasts (MEFs) from telomerase- and p53-deficient mice demonstrate that p53 mediates the adverse effects of critically short telomeres (44). Coincident with severe telomere shortening and associated genetic instability, p53 is activated, thereby leading to growth arrest and/or apoptosis (45). Progressive telomere shortening in oral keratinocytes may induce the p53 and likely the pRb pathways. High levels of p16 expression in presenescent OKF6-D1 cells in our study support this notion, as it has been shown that senescence is accompanied by increased p16 expression (4, 46). Disruption of p53 function in concert with cyclin D1 overexpression reproducibly enables oral keratinocytes to overcome senescence, thereby leading to immortalization despite maintained p16 levels.
Telomere shortening in cells from telomerase-deficient mice results in loss of telomere function and consequently genetic instability with chromosomal end-to-end fusions (47). Cells with short telomeres and intact p53 response may be efficiently eliminated from the culture by apoptosis or growth arrest. Consequently, cells from telomerase-deficient and p53–/– or p53+/– mice display increased chromosomal instability (44, 48). Although these mice may not represent the ideal model for human carcinogenesis owing to the differences in telomere biology (49, 50), it is compelling that mice deficient for telomerase and p53, which acquire genetic instability and force cells to use ALT, notably develop squamous epithelial cancers among other tumor types (48). By analogy, disruption of p53 function in combination with cyclin D1 overexpression in human oral keratinocytes induces the same type of genetic instability, including end-to-end fusions and translocations. These chromosomal rearrangements are consistent with the selection of chromosomal changes in cells containing critically short telomeres, which could also have led to the selection of ALT.
Surprisingly, we did not detect widespread cell death or apoptosis characteristic of crisis during the passage of OKF6-D1/Δp53 cells. Instead, we observed that telomere length shortened in these cells until PD 120. We subsequently observed striking telomere lengthening by PD 160. Although we did not detect evidence for crisis, karyotype analysis of these immortal cells showed clear evidence of end-to-end fusions often seen in cells that have survived crisis. We suspect that the expression of cyclin D1 and dominant-negative p53 permitted the activation of ALT at high frequency in our cultures; however, we cannot yet determine whether the expression of these genes merely created a milieu that fosters the activation of ALT or if it directly induced ALT. Importantly, although ALT cell lines have been derived from cells expressing SV40 large T antigen, the immortalized cells presented here are the first of human epithelial origin to have been generated using genes known to be altered in human cancers. This implies that similar mechanisms may operate during squamous cell carcinogenesis.
In summary, overexpression of cyclin D1 together with a dominant-negative form of p53 led to the immortalization of oral keratinocytes at high frequency through a telomerase-independent ALT mechanism. This system represents the first model in which ALT was generated using genetic alterations commonly observed in human cancers. We anticipate that this model system will enable us to understand not only the mechanism of ALT activation but also to delineate further the discrete steps that lead to squamous cell malignant transformation.
Acknowledgments
This work was supported by grants from the NIH (R01-DK53377 and P01-DE12467 to A.K. Rustgi), by the Abramson Family Cancer Research Institute (A.K. Rustgi), by grants from Deutsche Krebshilfe (D/96/17197 and 10-1656-Op 1 to O.G. Opitz), by the Howard Hughes Medical Institute Postdoctoral Fellowship (W.C. Hahn), and by a Doris Duke Charitable Foundation Clinical Scientist Development Award (W.C. Hahn). We are grateful to Steven Reeves for the cyclin D1 retroviral expression vector; to Ed Harlow for cyclin D1 and p16 antibodies; and to Hiroshi Nakagawa, Felix Brembeck, and Jun-ichi Okano for discussions. Gita Goessel provided excellent technical assistance. We are also grateful for the advice and support of Robert A. Weinberg.
Footnotes
See the related Commentary beginning on page 665.
References
- 1.Ness SL, et al. Mouse keratin 4 is necessary for internal epithelial integrity. J Biol Chem. 1998;273:23904–23911. doi: 10.1074/jbc.273.37.23904. [DOI] [PubMed] [Google Scholar]
- 2.Opitz OG, Jenkins TD, Rustgi AK. Transcriptional regulation of the differentiation-linked human K4 promoter is dependent upon esophageal-specific nuclear factors. J Biol Chem. 1998;273:23912–23921. doi: 10.1074/jbc.273.37.23912. [DOI] [PubMed] [Google Scholar]
- 3.Hayflick L. The limited in vitro lifespan of human diploid cell strains. Exp Cell Res. 1965;37:614–621. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Loughran O, et al. Evidence for the inactivation of multiple replicative lifespan genes in immortal human squamous cell carcinoma keratinocytes. Oncogene. 1997;14:1955–1964. doi: 10.1038/sj.onc.1201028. [DOI] [PubMed] [Google Scholar]
- 5.Allsopp RC, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddel RR. The role of senescence and immortalization in carcinogenesis. Carcinogenesis. 2000;21:477–484. doi: 10.1093/carcin/21.3.477. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez RD, et al. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harley CB, Futcher AB, Greider CW. Telomeres shorten during aging of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 10.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 11.Califano J, et al. Detection of telomerase activity in oral rinses from head and neck squamous cell carcinoma patients. Cancer Res. 1996;56:5720–5722. [PubMed] [Google Scholar]
- 12.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 13.Kiyono T, et al. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 14.Dickson MA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)- enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright WE, Pereira-Smith OM, Shay JW. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol Cell Biol. 1989;9:3088–3092. doi: 10.1128/mcb.9.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryan TM, Reddel RR. SV40-induced immortalization of human cells. Crit Rev Oncog. 1994;5:331–357. doi: 10.1615/critrevoncog.v5.i4.10. [DOI] [PubMed] [Google Scholar]
- 18.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 19.Bartkova J, et al. Abnormal patterns of D-type cyclin expression and G1 regulation in human head and neck cancer. Cancer Res. 1995;55:949–956. [PubMed] [Google Scholar]
- 20.Nakagawa H, et al. Human cyclin D1 oncogene and esophageal squamous cell carcinoma. Cancer. 1995;76:541–549. doi: 10.1002/1097-0142(19950815)76:4<541::aid-cncr2820760402>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa H, et al. The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach. Oncogene. 1997;14:1185–1190. doi: 10.1038/sj.onc.1200937. [DOI] [PubMed] [Google Scholar]
- 22.Wong DT, Todd R, Tsuji T, Donoff RB. Molecular biology of human oral cancer. Crit Rev Oral Biol Med. 1996;7:319–328. doi: 10.1177/10454411960070040201. [DOI] [PubMed] [Google Scholar]
- 23.Lindberg K, Rheinwald JG. Three distinct keratinocyte subtypes identified in human oral epithelium by their patterns of keratin expression in culture and in xenografts. Differentiation. 1990;45:230–241. doi: 10.1111/j.1432-0436.1990.tb00477.x. [DOI] [PubMed] [Google Scholar]
- 24.Hiyama H, Iavarone A, LaBaer J, Reeves SA. Regulated ectopic expression of cyclin D1 induces transcriptional activation of the cdk inhibitor p21 gene without altering cell cycle progression. Oncogene. 1997;14:2533–2542. doi: 10.1038/sj.onc.1201080. [DOI] [PubMed] [Google Scholar]
- 25.Opitz OG, Rustgi AK. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 2000;60:2825–2830. [PubMed] [Google Scholar]
- 26.Thompson DA, et al. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene. 1997;15:3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- 27.Hahn WC, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 28.Harris CC. p53: at the crossroads of molecular carcinogenesis and risk assessment. Science. 1993;262:1980–1981. doi: 10.1126/science.8266092. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 30.Bova RJ, et al. Cyclin D1 and p16INK4A expression predict reduced survival in carcinoma of the anterior tongue. Clin Cancer Res. 1999;5:2810–2819. [PubMed] [Google Scholar]
- 31.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee RJ, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20:672–683. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovec H, Sewing A, Lucibello FC, Muller R, Moroy T. Oncogenic activity of cyclin D1 revealed through cooperation with Ha-ras: link between cell cycle control and malignant transformation. Oncogene. 1994;9:323–326. [PubMed] [Google Scholar]
- 34.Nylander K, Dabelsteen E, Hall PA. The p53 molecule and its prognostic role in squamous cell carcinomas of the head and neck. J Oral Pathol Med. 2000;29:413–425. doi: 10.1034/j.1600-0714.2000.290901.x. [DOI] [PubMed] [Google Scholar]
- 35.Bond JA, Wyllie FS, Wynford-Thomas D. Escape from senescence in human diploid fibroblasts induced directly by mutant p53. Oncogene. 1994;9:1885–1889. [PubMed] [Google Scholar]
- 36.Rogan EM, et al. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol Cell Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 38.Counter CM, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 40.Bryan TM, Reddel RR. Telomere dynamics and telomerase activity in in vitro immortalised human cells. Eur J Cancer. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 41.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 42.McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 43.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 44.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 45.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 46.Dai CY, Enders GH. p16 INK4a can initiate an autonomous senescence program. Oncogene. 2000;19:1613–1622. doi: 10.1038/sj.onc.1203438. [DOI] [PubMed] [Google Scholar]
- 47.Hande MP, Samper E, Lansdorp P, Blasco MA. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 49.Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 50.Artandi SE, DePinho RA. Mice without telomerase: what can they teach us about human cancer? Nat Med. 2000;6:852–855. doi: 10.1038/78595. [DOI] [PubMed] [Google Scholar]