Abstract
Background
We studied linkage disequilibrium (LD) patterns at the BRCA1 locus, a susceptibility gene for breast and ovarian cancer, using a dense set of 114 single nucleotide polymorphisms in 5 population groups. We focused on Ashkenazi Jews in whom there are known founder mutations, to address the question of whether we would have been able to identify the 185delAG mutation in a case-control association study (should one have been done) using anonymous genetic markers. This mutation is present in approximately 1% of the general Ashkenazi population and 4% of Ashkenazi breast cancer cases. We evaluated LD using pairwise and haplotype-based methods, and assessed correlation of SNPs with the founder mutations using Pearson's correlation coefficient.
Results
BRCA1 is characterized by very high linkage disequilibrium in all populations spanning several hundred kilobases. Overall, haplotype blocks and pair-wise LD bins were highly correlated, with lower LD in African versus non-African populations. The 185delAG and 5382insC founder mutations occur on the two most common haplotypes among Ashkenazim. Because these mutations are rare, even though they are in strong LD with many other SNPs in the region as measured by D-prime, there were no strong associations when assessed by Pearson's correlation coefficient, r (maximum of 0.04 for the 185delAG).
Conclusion
Since the required sample size is related to the inverse of r, this suggests that it would have been difficult to map BRCA1 in an Ashkenazi case-unrelated control association study using anonymous markers that were linked to the founder mutations.
Background
Numerous advances in our understanding of genetic susceptibility to breast cancer have been made over the past decade, most notably the discovery of BRCA1 in 1994 and BRCA2 in 1995 [1,2]. Mutations in these genes account for approximately 2/3 of families with clearly inherited forms of breast and ovarian cancer (5 or more cases in a family) [3,4]. In addition to the high-penetrance genes BRCA1/BRCA2, rare mutations in a number of other genes, such as CHEK2, ATM, BRIP1, and PALB1 predispose to breast cancer [5-9], as do more common variants in CASP8 and TGFB1[10]. The total heritability of breast cancer is at least 10% [11,12], and possibly up to 25% or higher [13,14]. Mutations in known high-risk genes, however, account for a relatively small proportion (probably less than 20%) of the excess risk due to genetic factors [15,16].
Fueled by the completion of the first phases of the HapMap project [17], which characterized common variation within the genome of four population groups, there is considerable interest in using these resources to map susceptibility genes for common, complex diseases. The genome-wide case-control association study, whereby the prevalence of genetic variants is compared between cases and unrelated control subjects without the disease, may have the greatest power to identify novel susceptibility genes [18-20]. They rely on using a very dense set of markers that capture a significant fraction of all common genetic variation, such that the variants assayed either include those that are biologically relevant, or those which are highly-correlated with the former due to linkage disequilibrium. Although some design and analysis issues remain, numerous common variants have now been identified for breast cancer and other conditions using this design [21,22].
Breast cancer may serve as a useful paradigm for common, complex disease mapping studies because, while a portion of susceptibility genes have been identified, the majority of the residual familial clustering remains unexplained, and is likely to be polygenic in nature, due to a number of lower-penetrance genes in the context of environmental exposures [23,24]. Furthermore, there are two common Ashkenazi Jewish (AJ) founder BRCA1 mutations, 185delAG and 5382insC, initially identified in linkage studies of multiple-case breast/ovarian cancer families [25]. This contrasts with most other populations in which there are numerous unique BRCA1/BRCA2 mutations, with none present at 1% or greater population frequency. The BRCA1 AJ founder mutations account for the majority of Jewish breast-ovarian cancer families, and are present in approximately 1% of the general Jewish population [26]. The AJ founder mutations, owing to their high prevalence compared to other populations, offered an opportunity to test whether they might have been identified through a case-control association study of the kind suggested as the new gene discovery strategy in the post-HapMap era.
Results
SNP allele frequencies
A total of 289 unrelated reference subjects selected without regard to breast cancer from five population groups (48 each from African-Americans, Chinese-Americans and Mexican-Americans, 60 CEPH subjects, and 85 Ashkenazi Jews) were genotyped across BRCA1, spanning a region of approximately 646 kb. Table 1 presents the allele frequencies and Hardy-Weinberg P-vales for all the 112 polymorphic SNPs and the two founder mutations that were typed for all 5 populations. Eight of 570 tests showed departures from equilibrium at the 0.01 level, but because none of the eight showed Mendelian segregation errors within families, and because of the number of comparisons performed, they were not excluded from later calculations. Allele frequencies were generally highly correlated among Ashkenazi Jews, CEPH, Chinese-Americans and Mexican-Americans (minimum r >0.82), whereas African-Americans presented the lowest correlation values with all the other populations (maximum r <0.44). The highest correlation was found between Ashkenazi Jews and CEPH (r = 0.947) and the lowest between African-Americans and Mexican-Americans (r = 0.362). Most of the private SNPs (n = 18) originated in the African-American samples, although private SNPs were also observed in Ashkenazi Jews (n = 4), Chinese-Americans (n = 3), and Mexican-Americans (n = 1) (Table 1). Total observed heterozygosity for each marker across the five populations ranged from 0.4% for private SNPs to 49.3% [see Additional file 1]. FST ranged from 0.0047 (SNP8 – rs8176072) to 0.4338 (SNP102 – rs2593595). Sixty three percent of the SNPs showed little genetic differentiation (FST < 0.05), followed by twenty eight percent with moderate (0.05–0.15) and less than ten percent with higher genetic differentiation. We also calculated pair-wise FST measures, and the distribution was very similar for the Ashkenazi Jews, CEPH, Chinese-Americans and Mexican-Americans versus each other (range from 0.008–0.018) as compared with African-Americans versus all the other populations (range from 0.082–0.092), showing that African-Americans had by far the greatest level of differentiation. These results are congruent with the low allele frequency correlation values observed between African-Americans versus all the other groups.
Table 1.
SNP Frequencies and Hardy-Weinberg P-Values
| Ashkenazi Jews (n = 85) | CEPH (n = 60) | African Americans (n = 48) | Chinese Americans (n = 48) | Mexican Americans (n = 48) | ||||||||||||
| rs number | SNP Number | SNP name | Position b33a | Position b36.1b | Com Allelec | Min Alleled | mafe | P-value | maf | P-value | maf | P-value | maf | P-value | maf | P-value |
| 13119 | 1 | C_9270420 | 41373486 | 38718247 | c | t | 0.424 | 0.580 | 0.350 | 0.712 | 0.177 | 0.624 | 0.396 | 0.359 | 0.458 | 0.961 |
| 748620 | 2 | C_9270421 | 41373037 | 38717798 | c | g | 0.440 | 0.640 | 0.350 | 0.712 | 0.240 | 0.550 | 0.391 | 0.518 | 0.465 | 0.669 |
| 17599948 | 3 | C_9270454 | 41364175 | 38708936 | a | g | 0.211 | 0.649 | 0.192 | 0.865 | 0.104 | 0.022 | 0.094 | 0.326 | 0.362 | 0.591 |
| 11653231 | 4 | C_95201 | 41240225 | 38584986 | g | a | 0.435 | 0.703 | 0.350 | 0.712 | 0.240 | 0.550 | 0.396 | 0.359 | 0.467 | 0.905 |
| 9908805 | 5 | C_3178692 | 41230675 | 38575436 | c | t | 0.008 | 0.948 | 0.375 | 0.878 | 0.011 | 0.941 | ||||
| 2175957 | 6 | C_11631183 | 41195587 | 38540348 | t | g | 0.447 | 0.665 | 0.350 | 0.712 | 0.250 | 0.441 | 0.396 | 0.359 | 0.446 | 0.935 |
| 3092986 | 7 | BR1_000340 | 41186761 | 38531522 | a | g | 0.051 | 0.636 | 0.042 | 0.736 | 0.010 | 0.942 | 0.031 | 0.823 | ||
| 8176072 | 8 | BR1_000392 | 41186709 | 38531470 | t | a | 0.006f | 0.957 | ||||||||
| 8176074 | 9 | BR1_000571 | 41186530 | 38531291 | g | a | 0.010 | 0.942 | ||||||||
| 3765640 | 10 | C_2615164 | 41185012 | 38529773 | a | g | 0.440 | 0.640 | 0.350 | 0.712 | 0.228 | 0.740 | 0.394 | 0.433 | 0.444 | 0.592 |
| NAg | 11 | M_185delAG | 41184811 | 38529572 | a | - | 0.006 | 0.957 | ||||||||
| 8176090 | 12 | BR1_007918 | 41179183 | 38523944 | c | g | 0.008 | 0.948 | 0.096 | 0.338 | ||||||
| 1800062 | 13 | BR1_010574 | 41176528 | 38521289 | g | a | 0.021 | 0.883 | ||||||||
| 8176101 | 14 | BR1_010796 | 41176306 | 38521067 | a | t | 0.021 | 0.882 | ||||||||
| 8176103 | 15 | C_2615171 | 41175815 | 38520576 | g | a | 0.463 | 0.291 | 0.350 | 0.712 | 0.234 | 0.640 | 0.396 | 0.359 | 0.458 | 0.961 |
| 8176104 | 16 | BR1_011340 | 41175762 | 38520523 | g | a | 0.024 | 0.824 | 0.033 | 0.789 | 0.010 | 0.942 | ||||
| 8176109 | 17 | C_2615172 | 41174541 | 38519302 | a | g | 0.441 | 0.497 | 0.350 | 0.712 | 0.239 | 0.609 | 0.396 | 0.359 | 0.469 | 0.751 |
| 8065872 | 18 | BR1_016775 | 41170328 | 38515089 | t | a | 0.135 | 0.278 | 0.010 | 0.942 | ||||||
| 8176120 | 19 | BR1_017105 | 41169998 | 38514759 | g | a | 0.424 | 0.913 | 0.350 | 0.712 | 0.240 | 0.550 | 0.396 | 0.359 | 0.458 | 0.961 |
| 799914 | 20 | BR1_018573 | 41168546 | 38513307 | g | a | 0.135 | 0.278 | ||||||||
| 799913 | 21 | BR1_019408 | 41167711 | 38512472 | a | g | 0.032 | 0.772 | 0.042 | 0.736 | 0.365 | 0.813 | 0.010 | 0.942 | ||
| 8176128 | 22 | BR1_019904 | 41167215 | 38511976 | t | a | 0.063 | 0.644 | ||||||||
| 8176133 | 23 | BR1_020896 | 41166223 | 38510984 | t | g | 0.422 | 0.738 | 0.350 | 0.712 | 0.167 | 0.488 | 0.396 | 0.359 | 0.458 | 0.961 |
| 799912 | 24 | C_2615180 | 41165899 | 38510660 | c | t | 0.472 | 0.701 | 0.364 | 0.926 | 0.125 | 0.322 | 0.396 | 0.359 | 0.489 | 0.882 |
| 799923 | 25 | BR1_026422 | 41160696 | 38505457 | c | t | 0.232 | 0.031 | 0.308 | 0.858 | 0.042 | 0.763 | 0.104 | 0.459 | ||
| 8176145 | 26 | BR1_029258 | 41157859 | 38502620 | t | c | 0.400 | 0.038 | 0.319 | 0.586 | 0.240 | 0.550 | 0.404 | 0.309 | 0.448 | 0.829 |
| 8176146 | 27 | BR1_029448 | 41157669 | 38502430 | c | t | 0.031 | 0.774 | 0.008 | 0.948 | ||||||
| 7503154 | 28 | BR1_030748 | 41156369 | 38501130 | t | g | 0.415 | 0.618 | 0.339 | 0.478 | 0.223 | 0.771 | 0.396 | 0.359 | 0.468 | 0.862 |
| 1799950 | 29 | BR1_031875 | 41155246 | 38500007 | a | g | 0.077 | 0.442 | 0.042 | 0.736 | 0.010 | 0.942 | 0.031 | 0.823 | ||
| 4986850 | 30 | BR1_032885 | 41154236 | 38498997 | g | a | 0.133 | 0.096 | 0.113 | 0.071 | 0.033 | 0.819 | 0.013 | 0.935 | ||
| 16940 | 31 | BR1_033119 | 41154002 | 38498763 | t | c | 0.422 | 0.429 | 0.350 | 0.712 | 0.177 | 0.624 | 0.396 | 0.359 | 0.448 | 0.829 |
| 799917 | 32 | BR1_033420 | 41153701 | 38498462 | c | t | 0.429 | 0.766 | 0.358 | 0.868 | 0.125 | 0.322 | 0.396 | 0.359 | 0.479 | 0.571 |
| 4986852 | 33 | BR1_003927 | 41153194 | 38497955 | g | a | 0.012 | 0.912 | 0.042 | 0.736 | 0.010 | 0.942 | ||||
| 2227945 | 34 | BR1_034226 | 41152895 | 38497656 | a | g | 0.052 | 0.703 | 0.010 | 0.942 | ||||||
| 16942 | 35 | BR1_034356 | 41152765 | 38497526 | a | g | 0.433 | 0.868 | 0.348 | 0.615 | 0.245 | 0.521 | 0.396 | 0.359 | 0.447 | 0.716 |
| 799916 | 36 | C_7530109 | 41151955 | 38496716 | t | g | 0.438 | 0.482 | 0.358 | 0.868 | 0.271 | 0.703 | 0.394 | 0.433 | 0.479 | 0.894 |
| 2070833 | 37 | BR1_035507 | 41151614 | 38496375 | c | a | 0.031 | 0.001 | 0.271 | 0.726 | 0.135 | 0.167 | ||||
| 2070834 | 38 | BR1_036077 | 41151050 | 38495811 | a | c | 0.435 | 0.626 | 0.342 | 0.568 | 0.250 | 1.000 | 0.396 | 0.359 | 0.438 | 0.634 |
| 8176158 | 39 | BR1_036793 | 41150334 | 38495095 | a | g | 0.418 | 0.393 | 0.353 | 0.665 | 0.174 | 0.532 | 0.396 | 0.359 | 0.458 | 0.961 |
| 8176160 | 40 | BR1_036859 | 41150268 | 38495029 | a | g | 0.435 | 0.626 | 0.350 | 0.712 | 0.240 | 0.550 | 0.396 | 0.359 | 0.458 | 0.961 |
| 8176166 | 41 | BR1_038085 | 41149042 | 38493803 | a | g | 0.196 | 0.391 | 0.150 | 0.172 | 0.104 | 0.022 | 0.120 | 0.632 | 0.344 | 0.395 |
| 8176174 | 42 | BR1_040350 | 41146777 | 38491538 | a | t | 0.063 | 0.644 | ||||||||
| 3950989 | 43 | C_3178696 | 41146718 | 38491479 | g | a | 0.441 | 0.566 | 0.348 | 0.615 | 0.234 | 0.640 | 0.396 | 0.359 | 0.457 | 0.923 |
| 8176175 | 44 | BR1_040669 | 41146458 | 38491220 | t | - | 0.006 | 0.956 | 0.073 | 0.586 | ||||||
| 8176177 | 45 | BR1_041288 | 41145840 | 38490601 | a | g | 0.021 | 0.883 | ||||||||
| 8176178 | 46 | BR1_041721 | 41145407 | 38490168 | a | g | 0.042 | 0.763 | ||||||||
| 1060915 | 47 | C_3178676 | 41143235 | 38487996 | a | g | 0.380 | 0.105 | 0.352 | 0.851 | 0.177 | 0.624 | 0.396 | 0.359 | 0.468 | 0.862 |
| 3737559 | 48 | C_3178677 | 41143069 | 38487830 | c | t | 0.124 | 0.481 | 0.067 | 0.580 | 0.083 | 0.208 | 0.043 | 0.761 | ||
| 8176187 | 49 | BR1_045154 | 41141974 | 38486735 | t | c | 0.010 | 0.942 | ||||||||
| 8176188 | 50 | BR1_045505 | 41141623 | 38486384 | t | g | 0.063 | 0.644 | ||||||||
| 6416927 | 51 | BR1_0046019 | 41141109 | 38485870 | g | c | 0.083 | 0.208 | ||||||||
| 8176198 | 52 | BR1_047826 | 41139302 | 38484063 | t | a | 0.422 | 0.429 | 0.362 | 0.427 | 0.348 | 0.114 | 0.426 | 0.761 | 0.479 | 0.571 |
| 8176199 | 53 | BR1_0477839 | 41139289 | 38484050 | a | c | 0.341 | 0.594 | 0.258 | 0.502 | 0.135 | 0.167 | 0.177 | 0.624 | 0.396 | 0.135 |
| 4239147 | 54 | BR1_048551 | 41138577 | 38483338 | t | c | 0.440 | 0.640 | 0.324 | 0.677 | 0.271 | 0.703 | 0.394 | 0.433 | 0.452 | 0.711 |
| 8176206 | 55 | BR1_050244 | 41136885 | 38481646 | a | g | 0.052 | 0.703 | 0.010 | 0.942 | ||||||
| 2236762 | 56 | C_11621042 | 41135440 | 38480201 | a | t | 0.435 | 0.626 | 0.358 | 0.868 | 0.271 | 0.703 | 0.406 | 0.581 | 0.479 | 0.894 |
| 1799966 | 57 | C_2615208 | 41131859 | 38476620 | t | c | 0.456 | 0.456 | 0.350 | 0.712 | 0.233 | 0.781 | 0.396 | 0.359 | 0.469 | 0.751 |
| 3092987 | 58 | BR1_055669 | 41131488 | 38476249 | a | g | 0.416 | 0.545 | 0.350 | 0.712 | 0.170 | 0.510 | 0.396 | 0.359 | 0.458 | 0.961 |
| 8176225 | 59 | BR1_056796 | 41130361 | 38475122 | g | t | 0.031 | 0.823 | ||||||||
| 8176232 | 60 | BR1_058369 | 41128788 | 38473549 | c | t | 0.013 | 0.936 | ||||||||
| 8176234 | 61 | BR1_058614 | 41128545 | 38473306 | a | g | 0.437 | 0.345 | 0.350 | 0.712 | 0.239 | 0.609 | 0.396 | 0.359 | 0.458 | 0.961 |
| 8176235 | 62 | BR1_058834 | 41128325 | 38473086 | g | a | 0.339 | 0.872 | 0.258 | 0.502 | 0.167 | 0.488 | 0.396 | 0.359 | 0.436 | 0.972 |
| 8176236 | 63 | BR1_059589 | 41127570 | 38472331 | t | c | 0.008 | 0.948 | 0.359 | 0.556 | 0.010 | 0.942 | ||||
| 8176240 | 64 | BR1_060022 | 41127137 | 38471898 | t | c | 0.063 | 0.644 | ||||||||
| 8176242 | 65 | BR1_060520 | 41126639 | 38471400 | g | a | 0.433 | 0.538 | 0.350 | 0.712 | 0.170 | 0.510 | 0.406 | 0.581 | 0.458 | 0.961 |
| 8176245 | 66 | BR1_061014 | 41126145 | 38470906 | t | c | 0.063 | 0.644 | ||||||||
| 3092994 | 67 | C_2615220 | 41124590 | 38469351 | c | t | 0.434 | 0.471 | 0.350 | 0.712 | 0.228 | 0.242 | 0.396 | 0.359 | 0.266 | 0.808 |
| 8176259 | 68 | BR1_062588 | 41124571 | 38469332 | t | - | 0.052 | 0.703 | ||||||||
| 8176265 | 69 | BR1_0064398 | 41122761 | 38467522 | g | a | 0.417 | 0.477 | 0.350 | 0.712 | 0.167 | 0.729 | 0.394 | 0.433 | 0.260 | 0.848 |
| 2187603 | 70 | BR1_064501 | 41122658 | 38467419 | g | a | 0.422 | 0.429 | 0.350 | 0.712 | 0.167 | 0.729 | 0.396 | 0.359 | 0.260 | 0.848 |
| 8176273 | 71 | BR1_066741 | 41120418 | 38465179 | t | c | 0.424 | 0.580 | 0.342 | 0.568 | 0.167 | 0.729 | 0.396 | 0.359 | 0.260 | 0.848 |
| 8176278 | 72 | BR1_067978 | 41119181 | 38463942 | a | g | 0.008 | 0.948 | 0.500 | 0.564 | 0.021 | 0.883 | ||||
| 8066171 | 73 | BR1_068063 | 41119096 | 38463857 | g | t | 0.146 | 0.237 | 0.010 | 0.942 | ||||||
| NA | 74 | M_5382insC | 41117845 | 38462606 | - | c | 0.006 | 0.957 | ||||||||
| 8176289 | 75 | C_2615230 | 41114821 | 38459582 | t | c | 0.441 | 0.566 | 0.342 | 0.568 | 0.223 | 0.258 | 0.396 | 0.359 | 0.266 | 0.808 |
| 8176293 | 76 | BR1_073023 | 41114136 | 38458897 | a | - | 0.031 | 0.823 | ||||||||
| 4793192 | 77 | BR1_074008 | 41113155 | 38457916 | a | g | 0.435 | 0.626 | 0.336 | 0.794 | 0.229 | 0.214 | 0.396 | 0.359 | 0.260 | 0.848 |
| 8176296 | 78 | BR1_074807 | 41112356 | 38457117 | a | g | 0.435 | 0.703 | 0.350 | 0.712 | 0.229 | 0.214 | 0.396 | 0.359 | 0.260 | 0.848 |
| 3092988 | 79 | C_2615238 | 41110467 | 38455228 | c | t | 0.424 | 0.580 | 0.350 | 0.712 | 0.170 | 0.709 | 0.396 | 0.359 | 0.260 | 0.848 |
| 8176303 | 80 | BR1_076933 | 41110230 | 38454991 | a | g | 0.010 | 0.942 | 0.042 | 0.763 | ||||||
| 8176305 | 81 | BR1_077034 | 41110129 | 38454890 | a | g | 0.094 | 0.753 | 0.092 | 0.442 | 0.021 | 0.883 | 0.010 | 0.942 | ||
| 8176307 | 82 | BR1_077328 | 41109835 | 38454596 | t | c | 0.063 | 0.644 | ||||||||
| 8068463 | 83 | BR1_079220 | 41107943 | 38452704 | c | t | 0.125 | 0.322 | ||||||||
| 8176313 | 84 | BR1_079396 | 41107767 | 38452528 | g | a | 0.006 | 0.957 | 0.017 | 0.896 | ||||||
| 8176316 | 85 | BR1_080570 | 41106594 | 38451355 | c | a | 0.021 | 0.883 | ||||||||
| 8176318 | 86 | BR1_081125 | 41106039 | 38450800 | g | t | 0.435 | 0.963 | 0.350 | 0.712 | 0.167 | 0.729 | 0.385 | 0.491 | 0.250 | 1.000 |
| 12516 | 87 | BR1_081990 | 41105173 | 38449934 | c | t | 0.435 | 0.626 | 0.350 | 0.712 | 0.229 | 0.214 | 0.406 | 0.581 | 0.260 | 0.848 |
| 8176320 | 88 | BR1_082035 | 41105128 | 38449889 | g | a | 0.006 | 0.957 | 0.033 | 0.789 | 0.010 | 0.942 | ||||
| 8176321 | 89 | BR1_082199 | 41104964 | 38449725 | g | a | 0.010 | 0.942 | ||||||||
| 8176323 | 90 | BR1_082687 | 41104476 | 38449237 | c | g | 0.429 | 0.766 | 0.350 | 0.712 | 0.229 | 0.214 | 0.396 | 0.359 | 0.260 | 0.848 |
| 7223952 | 91 | C_11621012 | 41103649 | 38448411 | t | c | 0.441 | 0.566 | 0.356 | 0.765 | 0.281 | 0.569 | 0.396 | 0.359 | 0.287 | 0.931 |
| 9911630 | 92 | C_3178665 | 41097108 | 38441868 | a | g | 0.422 | 0.429 | 0.364 | 0.926 | 0.266 | 0.808 | 0.396 | 0.359 | 0.283 | 0.813 |
| 11460963 | 93 | C_2615245 | 41090062 | 38435121 | - | g | 0.441 | 0.497 | 0.350 | 0.712 | 0.228 | 0.242 | 0.396 | 0.359 | 0.266 | 0.808 |
| 2298861 | 94 | C_3178699 | 41085596 | 38430357 | g | a | 0.441 | 0.309 | 0.350 | 0.712 | 0.223 | 0.258 | 0.396 | 0.359 | 0.266 | 0.808 |
| 2298862 | 95 | C_3178698 | 41085453 | 38430214 | t | c | 0.447 | 0.383 | 0.350 | 0.712 | 0.208 | 0.343 | 0.396 | 0.359 | 0.260 | 0.848 |
| 443759 | 96 | C_2287905 | 41074499 | 38419260 | c | t | 0.229 | 0.129 | 0.241 | 0.656 | 0.354 | 0.520 | 0.083 | 0.208 | 0.146 | 0.981 |
| 11871636 | 97 | C_11617231 | 41063582 | 38408343 | a | c | 0.235 | 0.304 | 0.259 | 0.386 | 0.302 | 0.345 | 0.167 | 0.083 | 0.135 | 0.278 |
| 2271539 | 98 | C_1588447 | 41059714 | 38404475 | a | g | 0.388 | 0.932 | 0.388 | 0.339 | 0.213 | 0.447 | 0.448 | 0.341 | 0.277 | 0.768 |
| 690971 | 99 | C_765227 | 41025330 | 38370091 | g | t | 0.281 | 0.885 | ||||||||
| 528854 | 100 | C_1588417 | 41006873 | 38351634 | a | g | 0.106 | 0.230 | 0.098 | 0.488 | 0.479 | 0.990 | 0.063 | 0.644 | ||
| 323495 | 101 | C_1588405 | 40983252 | 38328013 | g | a | 0.316 | 0.182 | 0.342 | 0.568 | 0.208 | 0.001 | 0.396 | 0.135 | 0.443 | 0.407 |
| 2593595 | 102 | C_3256885 | 40965010 | 38309771 | a | g | 0.124 | 0.007 | 0.054 | 0.672 | 0.156 | 0.002 | 0.135 | 0.883 | 0.177 | <.0001 |
| 324075 | 103 | C_3256881 | 40935288 | 38280049 | a | g | 0.171 | 0.717 | 0.195 | 0.063 | 0.115 | 0.370 | 0.156 | 0.365 | 0.330 | 0.558 |
| 2290041 | 104 | C_15883310 | 40856085 | 38200846 | c | t | 0.012 | 0.913 | 0.302 | 0.103 | 0.031 | 0.823 | ||||
| 1078523 | 105 | C_2160077 | 40821525 | 38166286 | a | g | 0.480 | 0.158 | 0.370 | 0.650 | 0.135 | 0.883 | 0.478 | 0.369 | 0.474 | 0.075 |
| 752313 | 106 | C_1075621 | 40810589 | 38155350 | t | c | 0.363 | 0.314 | 0.422 | 0.851 | 0.292 | 0.954 | 0.406 | 0.250 | 0.351 | 0.251 |
| 7359598 | 107 | C_3256867 | 40806234 | 38150996 | c | t | 0.386 | 0.073 | 0.440 | 0.674 | 0.135 | 0.883 | 0.438 | 0.486 | 0.383 | 0.243 |
| 2271027 | 108 | C_15959277 | 40779567 | 38124328 | c | t | 0.065 | 0.636 | 0.031 | 0.823 | ||||||
| 7214055 | 109 | C_1441435 | 40761936 | 38106697 | c | g | 0.149 | 0.109 | 0.008 | 0.948 | 0.385 | 0.003 | 0.031 | 0.823 | 0.021 | 0.882 |
| 9766 | 110 | C_1441436 | 40761606 | 38106367 | g | a | 0.353 | 0.219 | 0.433 | 0.700 | 0.292 | 0.954 | 0.406 | 0.250 | 0.344 | 0.823 |
| 1553469 | 111 | C_7529639 | 40751527 | 38096288 | a | c | 0.065 | 0.248 | 0.058 | 0.061 | 0.031 | <.0001 | 0.202 | 0.943 | ||
| 2271029 | 112 | C_1125369 | 40744687 | 38089448 | c | a | 0.405 | 0.009 | 0.350 | 0.444 | 0.375 | 0.441 | 0.385 | 0.937 | 0.402 | 0.117 |
| 3760384 | 113 | C_1441438 | 40744216 | 38088977 | a | c | 0.359 | 0.020 | 0.392 | 0.666 | 0.447 | 0.716 | 0.385 | 0.937 | 0.458 | 0.023 |
| 2292749 | 114 | C_1441444 | 40727349 | 38072110 | c | t | 0.217 | 0.219 | 0.383 | 0.518 | 0.192 | 0.794 | 0.375 | 0.878 | 0.128 | 0.316 |
| Number of polymorphic loci | 83 | 80 | 99 | 71 | 82 | |||||||||||
| Number of SNPs with maf<0.05 | 11 | 13 | 14 | 6 | 17 | |||||||||||
| Mean maf | 0.305 | 0.269 | 0.181 | 0.327 | 0.287 | |||||||||||
a April 2003 (Build 33) Position
b March 2006 (Build 36.1) Position
c Com Allele = Common allele
d Min Allele = Minor allele
e maf = Minor allele frequency
f Population private SNPs are shown underlined
gNA= Not Applicable – not in dbSNP
LD structure
In order to analyze the LD structure at the BRCA1 locus, we chose two methods that rely on different premises. The first is haplotype block analysis which identifies sequential and non-overlapping sets of variants in high LD, separated by low levels of LD that are consistent with historical recombination. In this method, all htSNPs need to be genotyped in order to capture most of the genetic variation [27]. The second is a binning method in which SNPs in one LD bin can be interleaved with SNPs in other overlapping bins. Under this approach, one TagSNP per bin needs to be tested in order to capture SNP diversity [28].
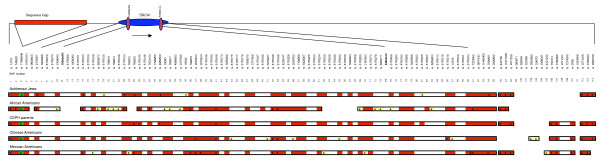
Our analyses of D' and r2 showed BRCA1 residing in a large region (~288 kb) of high LD (Fig. 1), in agreement with other reports [29-31]. The entire region studied showed long-range LD, falling primarily into three blocks among non-African populations. The block containing BRCA1 includes 95 SNPs and overlaps the largest LDSelect bin of SNPs correlated at r2 > 0.8 (Fig. 2 and Fig. 3).
Figure 1.
Comparison of haplotype blocks at 114 loci across five populations. Blocks were defined as in [27]; markers with MAF <0.05 are shown with a white background and were ignored in the calculations and block boundary estimation. Haplotype tag SNPs (htSNPs) within a block are indicated by arrowheads; htSNPs in only one population are shown on a yellow background while the single htSNP shared between all populations is shown on a green background.
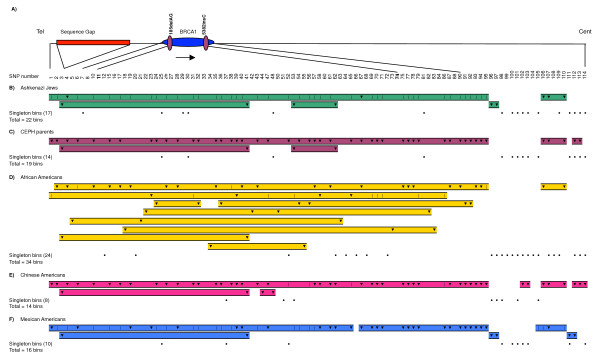
Figure 2.
Comparison of SNP bins derived from pair-wise measurements of linkage disequilibrium using LDSelect-Comp. SNPs with MAF < 5% do not have a vertical line or arrowhead in the column. A) Scale representation of the ~650 kb region studied, indicating the BRCA1 gene, founder mutations, and genome sequence gap of unknown true size. Anchor lines link to position of the SNP within the region. B-F) LDSelect creates bins of SNPs that have an r2 value of 0.8 or greater with at least one other SNP in the bin. Each vertical line and arrowhead represents a SNP, with dashed lines and shaded background connecting SNPs within the same bin. Down arrowheads indicate Tag SNPs (those with r2 ≥ 0.8 with all other SNPs in a bin). Note that this use of the term Tag-SNP is different from Haploview – with LDSelect, only one Tag-SNP per bin would be required to capture the majority of the nucleotide diversity. Singleton bins (SNPs that did not have r2 ≥ 0.8 with any other SNP) are indicated by solid dots on a single row. SNP number refers to numbering in column 1 of Table 1.
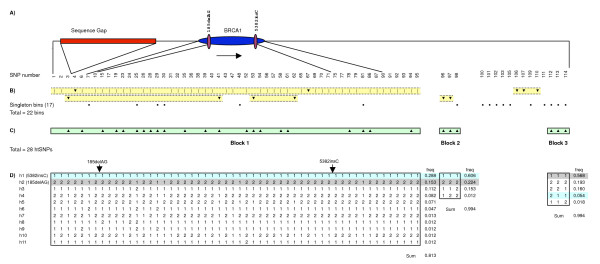
Figure 3.
Pair-wise measures of linkage disequilibrium and the two founder mutation-containing haplotypes. SNP number refers to numbering in column 1 of Table 1; only the 70 with MAF ≥ 0.05 in Ashkenazi Jews are shown in B-D. A) Scale representation of the ~650 kb region studied, indicating the BRCA1 gene, founder mutations, and genome sequence gap of unknown true size. B) LDSelect-Comp output showing a total of 22 bins for Ashkenazi Jews, with 17 "singleton" bins indicated by solid dots on a single row. C) Haploview output showing three block structures and related ht-SNPs (indicated with up arrowheads). D) Haplotypes estimated for 85 unrelated Ashkenazi Jews using SNPHAP as implemented in HapScope. The block boundaries were calculated in Haploview and overlaid on this figure. All haplotypes with an estimated frequency of at least 1% are displayed (h1 to h11), with individual frequencies and sums indicated to the right of the blocks. The common allele is designated "1" and the minor allele "2". The 185delAG and 5382insC containing haplotypes, determined from the family based genotypes, are indicated with gray (haplotype 2) and blue background (haplotype 1), respectively. Black arrows indicate the relative position of these two founder mutations.
African-Americans presented the least LD of all populations, with the presence of more distinct blocks within the region (Fig. 1). Maps for all five populations shared a break-point that maps approximately 20 kb downstream of the BRCA1 gene, between SNP95 (rs2298862) and SNP96 (rs443759). Among non-African groups, only Mexican-Americans exhibited an additional break point within the 288 kb block structure that encompasses BRCA1. The 3'end of the entire region showed less extensive LD but a similar pattern across all the groups. Only one htSNP (SNP3 – rs17599948) was found to be completely shared across populations, which is not unexpected since htSNPs often are population-specific [28] (Fig. 1).
When the bin-based approach was used, we found that bins were largely shared across different ethnic groups (Fig. 2). The differences across populations were related to the number of bins as well as the number and position of TagSNPs. As expected, African-Americans were the most diverse group, containing the highest number of bins (34), followed by Ashkenazi Jews (22), CEPH (19), Mexican-Americans and Chinese-American (14). This contrasts with 28 htSNPs in African-Americans, and 16, 13, 25 and 18 among Ashkenazi Jews, CEPH, Mexican- Americans and Chinese-Americans, respectively. Three TagSNPs were shared by all populations (SNP3 – rs17599948, SNP41 – rs8176166, and SNP67 – rs3092994), showing average MAF of 0.193, 0.183 and 0.335, respectively (Fig. 2). Mexican-Americans showed two disjoint bins of highly-correlated SNPs, rather than one extended bin structure as evidenced in Ashkenazim, CEPH and Chinese-Americans. The disjoint occurred between positions 38,471,400 (SNP65 – rs8176242) and 38,469,351 (SNP67 – rs3092994), mapping between introns 17 and 18 of BRCA1 (Fig. 2F). Interestingly, our results resemble what others have observed [32] in Native- Americans, namely an historical recombination event between introns 15 and 18. All five populations showed a large bin spanning ~288 kb encompassing SNP1 (rs13119) through SNP95 (rs2298862) (Fig. 2A–F), which represented the same extended region found in the block analysis. This large bin had 0.278 average MAF across populations and included BRCA1 coding polymorphisms L771L_(TTG>CTG), P871L_(CCG>CTG), 1183R_(AAA>AGA) and S1436S_(TCT>TCC) (Fig. 2).
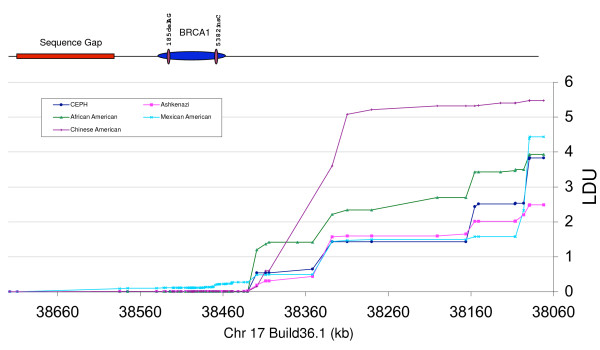
The maps of linkage disequilibrium in LD units (LDU) corresponded well with the two previous approaches of assessing disequilibrium. The four major breakpoints that were observed in Fig. 1 and 2, when haplotype blocks and bin structures were inferred, coincided with the same major steps in the LDU analysis (Fig. 4). In addition, we were able to observe two small steps in Fig. 4 for the Mexican-Americans, which were not observed in any other population. The first step occurred between SNP65 (rs8176242, intron 17) and SNP69 (rs8176265, intron 19). The corresponding site of possible recombination could be observed as a split at the main bin structure for Mexican-Americans in Fig. 2F. The second step was found between SNP90 (rs8176323) and SNP91 (rs7223952), downstream of the gene. A close correspondence was evidenced as a breakdown in LD around the same position in Fig. 1.
Figure 4.
LDU maps. Comparison of LDU maps across the ~650 kb region containing the BRCA1 gene for 5 populations. Top. Scale representation of the ~650 kb region studied, indicating the BRCA1 gene, founder mutations, and genome sequence gap of unknown true size. Bottom. LDU maps for five populations.
185delAG and 5382insC haplotype reconstruction
We were particularly interested in fine mapping the BRCA1 locus to identify possible gene variants or haplotypes associated with the two founder mutations in Ashkenazi Jews. Therefore, 82 intragenic and 30 flanking SNPs were tested against the two founder mutations with the Pearson's correlation (r) coefficient (Table 2). Although in strong LD with the majority of markers as measured by D' (Table 2), the highest pair-wise r2 for 185delAG was 0.04, owing to its relatively low frequency, with a common SNP (SNP96, rs443759) that mapped outside the large BRCA1-containing block, approximately 110 kb downstream of the gene. Regarding 5382insC, there was one highly-significant association (r2 = 1.0) with SNP8 (rs8176072) (Table 2). This is a rare SNP that was present only in 5382insC mutation carriers, and 5382insC was not correlated above 0.03 with any other SNP in the region.
Table 2.
Pair-wise correlation coefficients between the two founder BRCA1 mutations and all other SNPs among Ashkenazi Jews
| Ashkenazim (n = 85) | Correlation (r 2) with | D' with | D' with | |||||||
| SNP name | rs number | SNP number | SNP description | mafa | Hetb | HW P-value | 185delAG | 5382insC | 185delAG | 5382insC |
| C_9270420 | 13119 | 1 | NBR1: UTR | 0.424 | 0.52 | 0.580 | 0.011 | 0.001 | 1.000 | 1.000 |
| C_9270421 | 748620 | 2 | NBR1: UTR | 0.440 | 0.52 | 0.640 | 0.016 | 0.000 | 1.000 | 1.000 |
| C_9270454 | 17599948 | 3 | NBR1: intron | 0.211 | 0.35 | 0.649 | 0.016 | 0.001 | 1.000 | 1.000 |
| C_95201 | 11653231 | 4 | NBR1: intron | 0.435 | 0.51 | 0.703 | 0.014 | 0.001 | 1.000 | 1.000 |
| C_3178692 | 9908805 | 5 | LBRCA1: UTR | 0.000 | 1.000 | 1.000 | ||||
| C_11631183 | 2175957 | 6 | NBR2: intron | 0.447 | 0.52 | 0.665 | 0.013 | 0.001 | 1.000 | 1.000 |
| BR1_000340 | 3092986 | 7 | NBR2: intron | 0.051 | 0.10 | 0.636 | 0.001 | 0.000 | 1.000 | 1.000 |
| BR1_000392 | 8176072 | 8 | NBR2:intron | 0.006c | 0.01 | 0.957 | 0.000 | 1.000 | 1.000 | 1.000 |
| BR1_000571 | 8176074 | 9 | NBR2:UTR | 0.000 | 1.000 | 1.000 | ||||
| C_2615164 | 3765640 | 10 | BRCA1:UTR | 0.440 | 0.52 | 0.640 | 0.015 | 0.000 | 1.000 | 1.000 |
| M_185delAG | NAd | 11 | BRCA1:exon 2 | 0.006 | 0.01 | 0.957 | NA | 0.000 | NA | 1.000 |
| BR1_007918 | 8176090 | 12 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_010574 | 1800062 | 13 | BRCA1: P_K38K_(AAG>AAA) | 0.000 | 1.000 | 1.000 | ||||
| BR1_010796 | 8176101 | 14 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| C_2615171 | 8176103 | 15 | BRCA1: intron | 0.463 | 0.56 | 0.291 | 0.014 | 0.000 | 1.000 | 1.000 |
| BR1_011340 | 8176104 | 16 | BRCA1: intron | 0.024 | 0.05 | 0.824 | 0.001 | 0.000 | 1.000 | 1.000 |
| C_2615172 | 8176109 | 17 | BRCA1: intron | 0.441 | 0.53 | 0.497 | 0.011 | 0.000 | 1.000 | 0.000 |
| BR1_016775 | 8065872 | 18 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_017105 | 8176120 | 19 | BRCA1: intron | 0.424 | 0.49 | 0.913 | 0.015 | 0.001 | 1.000 | 1.000 |
| BR1_018573 | 799914 | 20 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_019408 | 799913 | 21 | BRCA1: intron | 0.032 | 0.06 | 0.772 | 0.001 | 0.000 | 1.000 | 1.000 |
| BR1_019904 | 8176128 | 22 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_020896 | 8176133 | 23 | BRCA1: intron | 0.422 | 0.51 | 0.738 | 0.012 | 0.000 | 1.000 | 1.000 |
| C_2615180 | 799912 | 24 | BRCA1: intron | 0.472 | 0.52 | 0.701 | 0.011 | 0.000 | 1.000 | 0.000 |
| BR1_026422 | 799923 | 25 | BRCA1: intron | 0.232 | 0.44 | 0.031 | 0.003 | 0.000 | 1.000 | 1.000 |
| BR1_029258 | 8176145 | 26 | BRCA1: intron | 0.400 | 0.59 | 0.038 | 0.021 | 0.001 | 1.000 | 0.000 |
| BR1_029448 | 8176146 | 27 | BRCA1: intron | 0.031 | 0.06 | 0.774 | 0.001 | 0.000 | 1.000 | 1.000 |
| BR1_030748 | 7503154 | 28 | BRCA1: intron | 0.415 | 0.51 | 0.618 | 0.012 | 0.001 | 1.000 | 1.000 |
| BR1_031875 | 1799950 | 29 | BRCA1: P_Q356R_(CAG>CGG) | 0.077 | 0.15 | 0.442 | 0.002 | 0.000 | 1.000 | 1.000 |
| BR1_032885 | 4986850 | 30 | BRCA1: P_D693N_(GAC>AAC) | 0.133 | 0.19 | 0.096 | 0.001 | 0.000 | 0.000 | 1.000 |
| BR1_033119 | 16940 | 31 | BRCA1: P_L771L_(TTG>CTG) | 0.422 | 0.53 | 0.429 | 0.012 | 0.001 | 1.000 | 1.000 |
| BR1_033420 | 799917 | 32 | BRCA1:P_P871L_(CCG>CTG) | 0.429 | 0.51 | 0.766 | 0.015 | 0.001 | 1.000 | 1.000 |
| BR1_003927 | 4986852 | 33 | BRCA1: P_S1040N_(AGC>AAC) | 0.012 | 0.02 | 0.912 | 0.000 | 0.000 | 1.000 | 1.000 |
| BR1_034226 | 2227945 | 34 | BRCA1: P_S1140G_(AGT>GGT) | 0.000 | 1.000 | 1.000 | ||||
| BR1_034356 | 16942 | 35 | BRCA1: P_K1183R_(AAA>AGA) | 0.433 | 0.50 | 0.868 | 0.015 | 0.001 | 1.000 | 1.000 |
| C_7530109 | 799916 | 36 | BRCA1: intron | 0.438 | 0.53 | 0.482 | 0.015 | 0.000 | 1.000 | 1.000 |
| BR1_035507 | 2070833 | 37 | BRCA1: intron | 0.031 | 0.04 | 0.001 | 0.001 | 0.000 | 1.000 | 1.000 |
| BR1_036077 | 2070834 | 38 | BRCA1: intron | 0.435 | 0.52 | 0.626 | 0.014 | 0.001 | 1.000 | 1.000 |
| BR1_036793 | 8176158 | 39 | BRCA1: intron | 0.418 | 0.54 | 0.393 | 0.012 | 0.000 | 1.000 | 1.000 |
| BR1_036859 | 8176160 | 40 | BRCA1: intron | 0.435 | 0.52 | 0.626 | 0.014 | 0.001 | 1.000 | 1.000 |
| BR1_038085 | 8176166 | 41 | BRCA1: intron | 0.196 | 0.35 | 0.391 | 0.018 | 0.000 | 0.388 | 1.000 |
| BR1_040350 | 8176174 | 42 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| C_3178696 | 3950989 | 43 | BRCA1: intron | 0.441 | 0.52 | 0.566 | 0.007 | 0.000 | 1.000 | 1.000 |
| BR1_040669 | 8176175 | 44 | BRCA1: intron | 0.006 | 0.01 | 0.956 | 0.000 | 0.000 | 1.000 | 1.000 |
| BR1_041288 | 8176177 | 45 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_041721 | 8176178 | 46 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| C_3178676 | 1060915 | 47 | BRCA1: P_S1436S_(TCT>TCC) | 0.380 | 0.56 | 0.105 | 0.018 | 0.001 | 1.000 | 0.000 |
| C_3178677 | 3737559 | 48 | BRCA1: intron | 0.124 | 0.20 | 0.481 | 0.001 | 0.001 | 0.462 | 1.000 |
| BR1_045154 | 8176187 | 49 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_045505 | 8176188 | 50 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_0046019 | 6416927 | 51 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_047826 | 8176198 | 52 | BRCA1: intron | 0.422 | 0.53 | 0.429 | 0.016 | 0.001 | 1.000 | 1.000 |
| BR1_0477839 | 8176199 | 53 | BRCA1: intron | 0.341 | 0.42 | 0.594 | 0.016 | 0.000 | 1.000 | 1.000 |
| BR1_048551 | 4239147 | 54 | BRCA1: intron | 0.440 | 0.52 | 0.640 | 0.011 | 0.001 | 1.000 | 1.000 |
| BR1_050244 | 8176206 | 55 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| C_11621042 | 2236762 | 56 | BRCA1: intron | 0.435 | 0.52 | 0.626 | 0.016 | 0.001 | 1.000 | 1.000 |
| C_2615208 | 1799966 | 57 | BRCA1: intron | 0.456 | 0.54 | 0.456 | 0.008 | 0.000 | 1.000 | 0.185 |
| BR1_055669 | 3092987 | 58 | BRCA1: intron | 0.416 | 0.52 | 0.545 | 0.013 | 0.002 | 1.000 | 1.000 |
| BR1_056796 | 8176225 | 59 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_058369 | 8176232 | 60 | BRCA1: intron | 0.013 | 0.03 | 0.936 | 0.000 | 0.000 | 1.000 | 1.000 |
| BR1_058614 | 8176234 | 61 | BRCA1: intron | 0.437 | 0.54 | 0.345 | 0.016 | 0.000 | 1.000 | 1.000 |
| BR1_058834 | 8176235 | 62 | BRCA1: intron | 0.339 | 0.44 | 0.872 | 0.017 | 0.000 | 1.000 | 1.000 |
| BR1_059589 | 8176236 | 63 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_060022 | 8176240 | 64 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_060520 | 8176242 | 65 | BRCA1: intron | 0.433 | 0.52 | 0.538 | 0.011 | 0.001 | 1.000 | 1.000 |
| BR1_061014 | 8176245 | 66 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| C_2615220 | 3092994 | 67 | BRCA1: intron | 0.434 | 0.53 | 0.471 | 0.012 | 0.000 | 1.000 | 0.000 |
| BR1_062588 | 8176259 | 68 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_0064398 | 8176265 | 69 | BRCA1: intron | 0.417 | 0.52 | 0.477 | 0.012 | 0.001 | 1.000 | 1.000 |
| BR1_064501 | 2187603 | 70 | BRCA1: intron | 0.422 | 0.53 | 0.429 | 0.009 | 0.001 | 1.000 | 1.000 |
| BR1_066741 | 8176273 | 71 | BRCA1: intron | 0.424 | 0.52 | 0.580 | 0.011 | 0.001 | 1.000 | 1.000 |
| BR1_067978 | 8176278 | 72 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_068063 | 8066171 | 73 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| M_5382insC | NA | 74 | BRCA1:exon 20 | 0.006 | 0.01 | 0.957 | 0.000 | NA | 1.000 | NA |
| C_2615230 | 8176289 | 75 | BRCA1: intron | 0.441 | 0.52 | 0.566 | 0.014 | 0.000 | 1.000 | 1.000 |
| BR1_073023 | 8176293 | 76 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_074008 | 4793192 | 77 | BRCA1: intron | 0.435 | 0.52 | 0.626 | 0.016 | 0.001 | 1.000 | 1.000 |
| BR1_074807 | 8176296 | 78 | BRCA1: intron | 0.435 | 0.51 | 0.703 | 0.015 | 0.000 | 1.000 | 1.000 |
| C_2615238 | 3092988 | 79 | BRCA1: intron | 0.424 | 0.52 | 0.580 | 0.015 | 0.001 | 1.000 | 1.000 |
| BR1_076933 | 8176303 | 80 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_077034 | 8176305 | 81 | BRCA1: intron | 0.094 | 0.16 | 0.753 | 0.002 | 0.001 | 1.000 | 1.000 |
| BR1_077328 | 8176307 | 82 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_079220 | 8068463 | 83 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_079396 | 8176313 | 84 | BRCA1: intron | 0.006 | 0.01 | 0.957 | 0.000 | 0.000 | 1.000 | 1.000 |
| BR1_080570 | 8176316 | 85 | BRCA1: intron | 0.000 | 1.000 | 1.000 | ||||
| BR1_081125 | 8176318 | 86 | BRCA1: UTR | 0.435 | 0.49 | 0.963 | 0.009 | 0.001 | 1.000 | 1.000 |
| BR1_081990 | 12516 | 87 | BRCA1: UTR | 0.435 | 0.52 | 0.626 | 0.014 | 0.001 | 1.000 | 1.000 |
| BR1_082035 | 8176320 | 88 | BRCA1: UTR | 0.006 | 0.01 | 0.957 | 0.000 | 0.000 | 1.000 | 1.000 |
| BR1_082199 | 8176321 | 89 | Intergenic | 0.000 | 1.000 | 1.000 | ||||
| BR1_082687 | 8176323 | 90 | Intergenic | 0.429 | 0.51 | 0.766 | 0.015 | 0.001 | 1.000 | 1.000 |
| C_11621012 | 7223952 | 91 | Intergenic | 0.441 | 0.52 | 0.566 | 0.014 | 0.000 | 1.000 | 1.000 |
| C_3178665 | 9911630 | 92 | Intergenic | 0.422 | 0.53 | 0.429 | 0.016 | 0.001 | 1.000 | 1.000 |
| C_2615245 | 11460963 | 93 | Intergenic | 0.441 | 0.53 | 0.497 | 0.014 | 0.001 | 1.000 | 1.000 |
| C_3178699 | 2298861 | 94 | ARHN: locus | 0.441 | 0.55 | 0.309 | 0.011 | 0.001 | 0.000 | 1.000 |
| C_3178698 | 2298862 | 95 | ARHN: locus | 0.447 | 0.54 | 0.383 | 0.011 | 0.001 | 1.000 | 1.000 |
| C_2287905 | 443759 | 96 | IFI35:intron | 0.229 | 0.41 | 0.129 | 0.040 | 0.002 | 1.000 | 1.000 |
| C_11617231 | 11871636 | 97 | RPL27: intron | 0.235 | 0.40 | 0.304 | 0.036 | 0.002 | 1.000 | 1.000 |
| C_1588447 | 2271539 | 98 | RPL27: intron | 0.388 | 0.47 | 0.932 | 0.023 | 0.003 | 1.000 | 1.000 |
| C_765227 | 690971 | 99 | MGC2744: intron | 0.000 | 1.000 | 1.000 | ||||
| C_1588417 | 528854 | 100 | MGC2744: intergenic | 0.106 | 0.16 | 0.230 | 0.000 | 0.001 | 1.000 | 1.000 |
| C_1588405 | 323495 | 101 | G6PC: intergenic | 0.316 | 0.37 | 0.182 | 0.004 | 0.000 | 0.205 | 1.000 |
| C_3256885 | 2593595 | 102 | G6PC: intron | 0.124 | 0.15 | 0.007 | 0.000 | 0.001 | 1.000 | 1.000 |
| C_3256881 | 324075 | 103 | Intergenic | 0.171 | 0.29 | 0.717 | 0.000 | 0.000 | 0.266 | 1.000 |
| C_15883310 | 2290041 | 104 | PRKWNK4: mis-sense | 0.012 | 0.02 | 0.913 | 0.005 | 0.000 | 1.000 | 1.000 |
| C_2160077 | 4321242 | 105 | RAMP2: intron | 0.480 | 0.58 | 0.158 | 0.003 | 0.000 | 1.000 | 1.000 |
| C_1075621 | 752313 | 106 | EZH1: intergenic | 0.363 | 0.40 | 0.314 | 0.005 | 0.000 | 1.000 | 1.000 |
| C_3256867 | 7359598 | 107 | EZH1: intergenic | 0.386 | 0.57 | 0.073 | 0.005 | 0.001 | 1.000 | 0.000 |
| C_15959277 | 2271027 | 108 | EZH1: intron | 0.000 | 1.000 | 1.000 | ||||
| C_1441435 | 7214055 | 109 | CNTNAP1: UTR | 0.149 | 0.30 | 0.109 | 0.001 | 0.001 | 1.000 | 1.000 |
| C_1441436 | 9766 | 110 | CNTNAP1: UTR | 0.353 | 0.52 | 0.219 | 0.007 | 0.000 | 0.173 | 1.000 |
| C_7529639 | 1553469 | 111 | CNTNAP1: silent | 0.065 | 0.11 | 0.248 | 0.002 | 0.000 | 0.463 | 1.000 |
| C_1125369 | 2271029 | 112 | CNTNAP1: silent | 0.405 | 0.62 | 0.009 | 0.000 | 0.001 | 0.040 | 1.000 |
| C_1441438 | 3760384 | 113 | CNTNAP1: intron | 0.359 | 0.58 | 0.020 | 0.005 | 0.000 | 1.000 | 1.000 |
| C_1441444 | 2292749 | 114 | TUBG2: intron | 0.217 | 0.39 | 0.219 | 0.006 | 0.001 | 1.000 | 1.000 |
a maf = Minor allele frequency
bHet = Heterozygosity
cPopulation private SNPs are shown underlined
dNA= Not Applicable
Haplotypes were estimated for the set of all SNPs with MAF ≥ 0.05. Haplotypes for the founder mutation containing chromosomes were unambiguously determined in at least one family across the entire region studied. The185delAG and 5382insC mutations occurred on the two most common haplotypes, representing 15% (haplotype 2) and 29% (haplotype 1) of the chromosomes, respectively, among Ashkenazi Jews (Fig. 3D). In the haplotype analyses, the 185delAG mutation occurred on a chromosome with the minor allele at most loci, and the 5382insC on a chromosome with the major allele at most loci (Fig. 3D). This pattern constitutes what has been previously described as "yin yang haplotypes", in which two high-frequency haplotypes have different alleles at most SNP sites [33].
Discussion
The primary objective of this study was to address the question of whether we could identify the Ashkenazi BRCA1 founder mutation 185delAG in a typical case-control association study, using anonymous genetic markers. The answer is no. The impact on the required sample size (S) needed if one studies a marker in LD with the true disease allele is related to the inverse of the square of their correlation coefficient (r), as in S = 1/r2 [34]. Almost none of the SNPs had a high correlation coefficient with either founder mutation (Table 2). Since most markers were more common than the founder mutations, this result is not surprising. However, our SNP selection strategy did not exclude low frequency SNPs. In fact, one of the SNPs identified in 3 of the 90 Polymorphism Discovery Resource subjects [35] was perfectly correlated with 5382insC. However, we did not observe this SNP in any of the four non-Ashkenazi reference populations. Based on our results, the sample size required to detect the 185delAG mutation in a breast cancer case-control study conducted in Ashkenazi women that did not directly test for the mutation, would be at least 25 times larger than one that measured the mutation directly, requiring on the order of 62,000 subjects. Based on pair-wise measurements, we conclude that it would have been extremely difficult to have mapped the two founder mutations using the case-control association methodology using common SNPs.
Association studies may also compare combinations of SNPs, or haplotypes, between cases and controls, and the founder mutations might have been discoverable if they occurred on uncommon haplotypes. Using relatively common SNPs (MAF ≥ 5%), like those on whole-genome SNP platforms, we found that the two mutations were present on haplotypes representing a polar pattern, termed yin-yang haplotypes [33]. These two haplotypes accounted together for a large percentage of the total chromosomes studied, independent of the population, ranging from 64% for the Chinese-Americans to 43% for the CEPH.
It is highly unlikely that the founder mutations could have been discovered owing to a difference in haplotype frequency between cases and controls largely because they occur on the two most common haplotypes. For example, consider a case-control study of Ashkenazi Jews with 500 cases and 500 controls. Among controls (1000 chromosomes), the distribution of BRCA1-containing haplotypes would be roughly as in Fig. 3D (i.e., there would have been 288 chromosomes with haplotype 1 and 153 with haplotype 2). Among cases, assuming 1% carried 5382insC and 4% carried 185delAG, there would be 5 additional haplotype 1 (total = 293) and 20 additional haplotype 2 (total = 173) chromosomes. These case-control contrasts, 293 vs. 288 (OR 1.02) and 173 vs. 153, would require extremely large sample sizes of over 30,000 subjects to detect either mutation with 80% statistical power. Conversely, in the more advantageous situation in which the 185delAG mutation by chance occurred on a rare haplotype (for example, haplotype 8), there would have been 32 such chromosomes in cases vs. 12 in controls requiring approximately 4,800 subjects for the same statistical power.
The BRCA1 locus is well known to have significant LD [29,30]. Nonetheless, we found a marked differentiation between African-Americans and non-African Americans in the haplotype block analysis. Compared with African-Americans, the non-African American populations had less haplotype diversity and more extensive LD (Fig. 1). The increased number of crossovers along the entire region for African-Americans probably reflects older evolutionary events. Our data conform to previous findings [27], describing higher haplotype diversity as well as less extensive LD in the Yoruban and African American samples than in European and Asian populations. When SNP "bins" derived from pair-wise measurements of LD were compared, we found a greater extent of LD boundaries being shared across the five different ethnical groups (Fig. 2). Ashkenazi Jews and the CEPH population had highly similar patterns of LD, independently of the type of analysis used to generate the LD structures (haplotype, or pair-wise bin methods) (Fig. 1 and Fig. 2). Overall, haplotype blocks and bins showed similar patterns, probably owing to the strong LD present overall in this region. The LDU analysis showed a remarkable overall similarity with the two previous methods that were used to analyze LD (Fig. 4). There were basically four major breakdowns in LD downstream to BRCA1 that were largely shared across populations. Nevertheless, African-Americans presented more recombination events than the other four populations, consistent with the smaller block sizes showed in Fig. 1.
Conclusion
In summary, our detailed analyses of 114 polymorphic SNPs in a 646 kb region around BRCA1 in Ashkenazi Jews and other populations confirmed a high level of linkage disequilibrium across nearly the entire region. In addition to 85 unrelated Ashkenazi Jews, we over-sampled carriers of the founder mutations 185delAG and 5382insC and their relatives to more precisely calculate correlations with other markers and to molecularly determine the mutation associated haplotypes (these subjects were not included in allele frequency estimates). This allowed us to assess the likelihood of discovering the founder mutations by virtue of their association with individual SNPs or haplotypes that one would assay in a breast cancer case-control study in Ashkenazi Jews. We did not observe a high correlation coefficient between any individual SNP likely to be included in a genome-wide anonymous scan and either founder mutation. Our findings suggest that a study at least 25X larger (60,000 subjects or more) would have been required if the mutations were not tested for directly. The two founder mutations occur on the two most common haplotypes, representing over 40% of the chromosomes, also suggesting that a haplotype-based analysis would not have been successful at detecting either of the underlying mutations. These results are influenced heavily by the relative rarity of the founder mutations, as reflected by high values for Lewontin's D' measures of LD but low correlation coefficients. Our results suggest caution in using genome-wide association studies with common SNPs for detecting uncommon, disease-causing mutations.
Methods
Subjects
Independent subjects included 85 unrelated Ashkenazi Jews, 60 European-Americans (Utah) from the CEPH (The Centre d'Etude du Polymorphisme Humain) family collection, and 48 each from African-Americans, Chinese-Americans and Mexican-Americans (Human Diversity Collection, Coriell Cell Repository, Camden, NJ). The 30 children of the 60 Utah CEPH subjects were also assayed to test for Mendelian errors. In addition, six unrelated BRCA1:185delAG and three unrelated BRCA1:5382insC founder mutation carriers and their relatives [36], identified through the National Cancer Institute's Cancer Family Registry, were included in the study in order to establish mutation-associated haplotypes from family data. Mutation-associated haplotypes were inferred through inspection of genotypes for all available first-degree relatives of mutation carriers. The Ashkenazi Jewish samples were obtained from anonymous control subjects from the National Laboratory for the Genetics of Israeli Populations at Tel-Aviv University [37].
Marker selection and genotyping
The 90 kb BRCA1 locus was previously re-sequenced in 90 individuals representing five major US ethnic/population groups from the Polymorphism Discovery Resource (PDR-90) [35], by the University of Washington as part of the Environmental Genome Project (EGP) [38]. Samples consisted of 24 European-, 24 African- 24 Asian-, 12 Mexican-, and six Native-Americans. The geographic origin of individual donors, however, is masked and may not be used to assign allele frequencies to specific sub-populations. Most of the 301 variants identified were SNPs. Genotyping all 301 variants at this locus in the current study was not necessary since many are highly correlated. We developed the following strategy to identify a reduced set of variants that still captured much of the diversity of the region. Using the EGP data on all 299 biallelic single nucleotide substitutions (i.e., no lower minor allele frequency cutoff), and using custom software, we calculated all pair-wise correlations (r2) and created "clusters", defined as groups of SNPs that were perfectly correlated with all others in the cluster. Our method is similar to LDSelect 1.0 [28] except that it required that all pair-wise correlations of SNPs in a cluster be 1.0 (i.e., complete LD). LDSelect is typically used with a threshold value of r2 of 0.8, and SNPs are clustered into "LD bins" if their pair-wise r2 is at or above this threshold value with at least one other SNP (but not all) in the bin. Using an r2 of 1.0 resulted in more clusters than a lower r2 threshold, increasing the number of SNPs assayed in this study.
Taqman 5'-nuclease assays were developed through Applied Biosystems (Foster City, CA) Assay-by-Design service after first filtering for repetitive, non-unique, and low-complexity sequence. We developed assays for all "singleton" SNPs (those that did not have pair-wise r2 values of 1.0 with another SNP). For the 59 clusters with two or more SNPs, we chose one SNP from each cluster of two, three and four SNPs, and for 9 clusters of five or more SNPs, we chose one fourth of them for assay development. In addition, we selected all (n = 43) commercially available Assay-on-Demand assays (Applied Biosystems, Foster City, CA) that mapped within approximately 200 kb upstream and 400 kb downstream of the BRCA1 locus. This SNP set represented almost all known variants (or ones highly correlated) at this locus.
Of the 143 resulting assays, three were excluded due to technical problems (poor clustering or more than one Mendelian error), and 28 were not polymorphic in our complete sample set, leaving 112 polymorphic SNPs in addition to the two founder mutations. There were 82 BRCA1 intragenic SNPs (approximately one SNP per 1 kb) and 30 SNPs that mapped to the region outside BRCA1 (approximately one SNP per 20 kb). The allelic discrimination assays were performed in 5 microliter reactions in 384-well plates according to manufacturer's recommendations. Data were analyzed with the allelic discrimination SDS 2.1 software on an ABI 7900HT (Applied Biosystems, Foster City, CA), with manual determination of genotype clusters [see Additional file 2].
Statistical analysis
Allelic frequency and chi-square goodness-of-fit tests for Hardy-Weinberg equilibrium (HWE) were calculated using SAS/Genetics 9.1 (SAS Institute, Inc., Cary, North Carolina). To assess the correlation between the two founder mutations and all other SNPs, we over-sampled mutation carriers and calculated a weighted Pearson's correlation coefficient using SAS 9.1. We also tested association by use of Tagger [39] operates in either pairwise or aggressive mode, and we used both approaches to examine association. Heterozygosity levels, as well as the variation in gene frequencies between populations by means of their FST (Wright's F-statistics) were calculated using POPGENE 1.31 [40].
Haplotypes and their frequencies were inferred from genotypes across the entire region for each population separately, using the software package SNPHAP 1.3 [41], as implemented in Hapscope [42], for loci with minor allele frequencies (MAF) > 5%. SNPHAP uses the expectation-maximization algorithm to calculate maximum likelihood estimates of haplotype frequencies from unphased genotype data.
In order to compare LD patterns across different populations, we employed two different analyses, the first based on partitioning SNPs into haplotype blocks [27] using Haploview [43] and the second based on "bins" of correlated SNPs not constrained to be adjacent to each other [28]. The binning method used a modified version of LDSelect 1.0 that calculates composite LD measures [44], without assuming that loci are in Hardy-Weinberg equilibrium. We used an r2 threshold of 0.8 for binning SNPs, and filtered SNPs with population-specific MAF ≤ 0.05.
LDSelect identifies tagSNPs, representing those SNPs in a bin that have r2 values at or above the threshold with all other SNPs in a bin. Only one tagSNP in each bin needs to be assayed to capture the majority of the SNP diversity. The block method employed by Haploview groups adjacent SNPs in strong LD, defined as those with one-sided upper 95% confidence bound on D' >0.98 and the lower bound >0.7. In this method, haplotype tag SNPs (htSNPs) represents the set of SNPs that must be assayed in each block to capture all haplotypes at 1% frequency in the population.
LD maps were constructed from genotype data with the software LDMAP [45]. LD maps are scaled in linkage disequilibrium units (LDU) and show (when plotted against the physical map) a pattern of plateaus (reflecting regions of low haplotype diversity and low recombination) and steps (representing regions of historical recombination events).
We genotyped related individuals from families segregating 185delAG and 5382insC founder mutations in order to reconstruct their haplotypes. The 185delAG- and 5382insC-containing haplotypes were unambiguously determined from analyzing the genotypes of all available family members. The frequencies of these mutation-containing haplotypes were determined from SNPHAP analyses of the five populations separately. Block boundaries were defined based on Haploview analyses and overlaid upon the SNPHAP results.
We estimated the required number of subjects to have 80% statistical power to identify the 185delAG mutation if tested directly in a case-control study to be approximately 2492 using EpiInfo 4.0 [46], assuming equal numbers of cases and controls, alpha of 0.0001, and heterozygous carrier frequencies of 0.6% for controls and 3.3% for cases.
Competing interests
The author(s) declares that there are no competing interests.
Authors' contributions
LMP performed laboratory and statistical analysis and drafted the manuscript. MAP and LRF performed laboratory analysis. WHR and JZ wrote custom software and performed statistical analysis. MHG provided biomaterials and critically revised the manuscript. KO and NAE participated in the study design and analysis. AC provided design assistance and performed statistical analysis. JPS conceived and designed the study and performed statistical analysis. All authors read and approved the final manuscript.
Supplementary Material
Summary of F-statistics and heterozygosity for all loci. Heterozygosity levels, as well as the variation in gene frequencies between populations by means of their FST (Wright's F-statistics).
Genotypes for all 114 polymorphic SNPs in 5 populations. Raw genotypes. Populations: (1) CEPH (includes related individuals), (2) African Americans, (3) Chinese Americans, (4) Mexican Americans and (5) Ashkenazi Jews.
Acknowledgments
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, Center for Cancer Research and the Division of Cancer Epidemiology and Genetics, National Cancer Institute, U.S. Department of Health and Human Services.
Contributor Information
Lutécia H Mateus Pereira, Email: luteciap@hotmail.com.
Marbin A Pineda, Email: pinedama@mail.nih.gov.
William H Rowe, Email: rowew@mail.nih.gov.
Libia R Fonseca, Email: lcaba003@fiu.edu.
Mark H Greene, Email: greenem@mail.nih.gov.
Kenneth Offit, Email: offitk@mskcc.org.
Nathan A Ellis, Email: naellis@uchicago.edu.
Jinghui Zhang, Email: jinghuiz@mail.nih.gov.
Andrew Collins, Email: A.R.Collins@soton.ac.uk.
Jeffery P Struewing, Email: struewij@mail.nih.gov.
References
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phrlps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone D, Bayer S, Wray C, Bogden R, Dayanath P, Ward J, Tonin P, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Szabo CI, King MC. Population genetics of BRCA1 and BRCA2. Am J Hum Genet. 1997;60:1013–1020. [PMC free article] [PubMed] [Google Scholar]
- Tonin P, Weber B, Offit K, Couch F, Rebbeck TR, Neuhausen S, Godwin AK, Daly M, Wagner-Costalos J, Berman D, Grana G, Fox E, Kane MF, Kolodner RD, Krainer M, Haber DA, Struewing JP, Warner E, Rosen B, Lerman C, Peshkin B, Norton L, Serova O, Foulkes WD, Lynch HT, Lenoir GM, Narod SA, Garber JE. Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat Med. 1996;2:1179–1183. doi: 10.1038/nm1196-1179. [DOI] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton D, McGuffog L, Thompson D, Dunning A, Tee L, Baynes C, Healey C, Pharoah P, Ponder B, Seal S, Barfoot R, Sodha N, Eeles R, Stratton M, Rahman N, Peto J, Spurdle AB, Chen X, Chenevix-Trench G, Hopper JL, Giles GG, McCredie MRE, Holli KSK, Kallioniemi O, Eerola H, Vahteristo P, Blomqvist C, Nevanlinna H, Kataja V, Mannermaa A, et al. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, Lissowska J, Brinton L, Peplonska B, Southey MC, Hopper JL, McCredie MR, Giles GG, Fletcher O, Johnson N, dos Santos Silva I, Gibson L, Bojesen SE, Nordestgaard BG, Axelsson CK, Torres D, Hamann U, Justenhoven C, Brauch H, Chang-Claude J, Kropp S, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Granstrom C, Czene K. Attributable risks for familial breast cancer by proband status and morphology: a nationwide epidemiologic study from Sweden. Int J Cancer. 2002;100:214–219. doi: 10.1002/ijc.10467. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Czene K. Attributable risks of familial cancer from the Family-Cancer Database. Cancer Epidemiol Biomarkers Prev. 2002;11:1638–1644. [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Risch N. The genetic epidemiology of cancer: Interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- Peto J, Collins N, Barfoot R, Seal S, Warren W, Rahman N, Easton DF, Evans C, Deacon J, Stratton MR. Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer. J Natl Cancer Inst. 1999;91:943–949. doi: 10.1093/jnci/91.11.943. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: Implications for design of association studies. Genet Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J, Ponder BJ, Easton DF. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struewing JP, Brody LC, Erdos MR, Kase RG, Giambarresi TR, Smith SA, Collins FS, Tucker MA. Detection of eight BRCA1 mutations in 10 breast/ovarian cancer families, including 1 family with male breast cancer. Am J Hum Genet. 1995;57:1–7. [PMC free article] [PubMed] [Google Scholar]
- Struewing JP, Abeliovich D, Peretz T, Avishai N, Kaback MM, Collins FS, Brody LC. The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat Genet. 1995;11:198–200. doi: 10.1038/ng1095-198. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnen PE, Wang PJ, Kimmel M, Chakraborty R, Nelson DL. Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res. 2002;12:1846–1853. doi: 10.1101/gr.483802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Barker DF. Evidence for effective suppression of recombination in the chromosome 17q21 segment spanning RNU2-BRCA1. Am J Hum Genet. 1999;64:1427–1439. doi: 10.1086/302358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Penney KL, Stram DO, Riley S, McKean-Cowdin R, Le Marchand L, Altshuler D, Haiman CA. A haplotype-based case-control study of BRCA1 and sporadic breast cancer risk. Cancer Res. 2005;65:7516–7522. doi: 10.1158/0008-5472.CAN-05-0132. [DOI] [PubMed] [Google Scholar]
- Kidd JR, Speed WC, Pakstis AJ, Kidd KK. A 100 kb block encompassing BRCA1. Am J Hum Genet. 2003;73:173. [Google Scholar]
- Zhang J, Rowe WL, Clark AG, Buetow KH. Genomewide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet. 2003;73:1073–1081. doi: 10.1086/379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Kramer JL, Velazquez IA, Chen BE, Rosenberg PS, Struewing JP, Greene MH. Prophylactic oophorectomy reduces breast cancer penetrance during prospective, long-term follow-up of BRCA1 mutation carriers. J Clin Oncol. 2005;23:8629–8635. doi: 10.1200/JCO.2005.02.9199. [DOI] [PubMed] [Google Scholar]
- National Laboratory for the Genetics of Israeli Populations at Tel-Aviv University http://www.tau.ac.il/medicine/NLGIP/nlgip.htm
- Environmental Genome Project, University of Washington http://egp.gs.washington.edu/data/brca1/
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- POPGENE: the user-friendly shareware for population genetic analysis http://www.ualberta.ca/~fyeh/index.htm
- SNPHAP: a program for estimating frequencies of large haplotypes of SNPs http://www-gene.cimr.cam.ac.uk/clayton/software/snphap.txt
- Zhang J, Rowe WL, Struewing JP, Buetow KH. HapScope: a software system for automated and visual analysis of functionally annotated haplotypes. Nucleic Acids Res. 2002;30:5213–5221. doi: 10.1093/nar/gkf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Weir BS. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. doi: 10.2307/2529947. [DOI] [PubMed] [Google Scholar]
- Maniatis N, Collins A, Xu CF, McCarthy LC, Hewett DR, Tapper W, Ennis S, Ke X, Morton NE. The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci USA. 2002;99:2228–2233. doi: 10.1073/pnas.042680999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EpiInfo Version 3.4.1 http://www.cdc.gov/epiinfo/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of F-statistics and heterozygosity for all loci. Heterozygosity levels, as well as the variation in gene frequencies between populations by means of their FST (Wright's F-statistics).
Genotypes for all 114 polymorphic SNPs in 5 populations. Raw genotypes. Populations: (1) CEPH (includes related individuals), (2) African Americans, (3) Chinese Americans, (4) Mexican Americans and (5) Ashkenazi Jews.